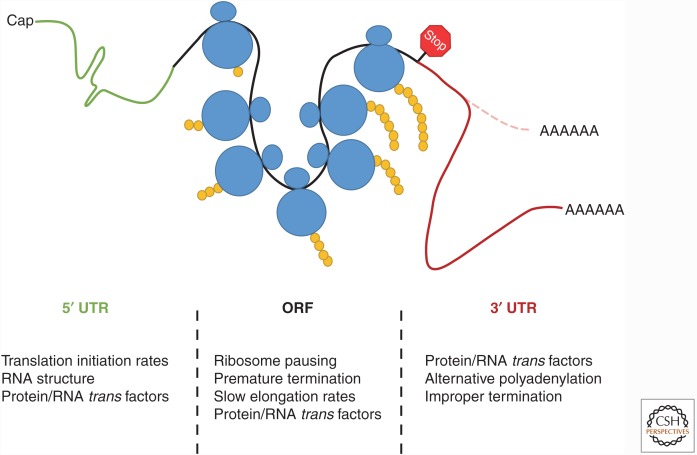
Figure 2.
Numerous regulatory points exist throughout the transcript to modulate co-translational messenger RNA (mRNA) decay. A brief overview of the factors and the location on the transcript that can influence the rate of mRNA decay in association with ribosome loading, elongation, and termination. In the 5′ untranslated region (UTR) (green), RNA structure, initiation rates, and RNA-binding protein (RBP)/RNA factors acting in trans can co-regulate mRNA decay and translation. As ribosomes (blue) read through the open reading frame (ORF) (black), elongation speed or lack thereof (ribosome pausing) is the primary feature that links active translation and mRNA decay. Events such as premature termination, RNA modification, or protein/RNA trans factor interaction can alter ribosomal speed, thus influencing mRNA stability. Finally, the 3′ UTR (red) can modulate mRNA decay and translation via interactions with other protein/RNA factors, different 3′ UTR isoforms that can arise by alternative polyadenylation (APA), or via improper termination, which can result from a defective ribosome or lack of a stop codon. The cell uses all of these regulatory mechanisms to ensure that accurate and efficient translation is achieved.