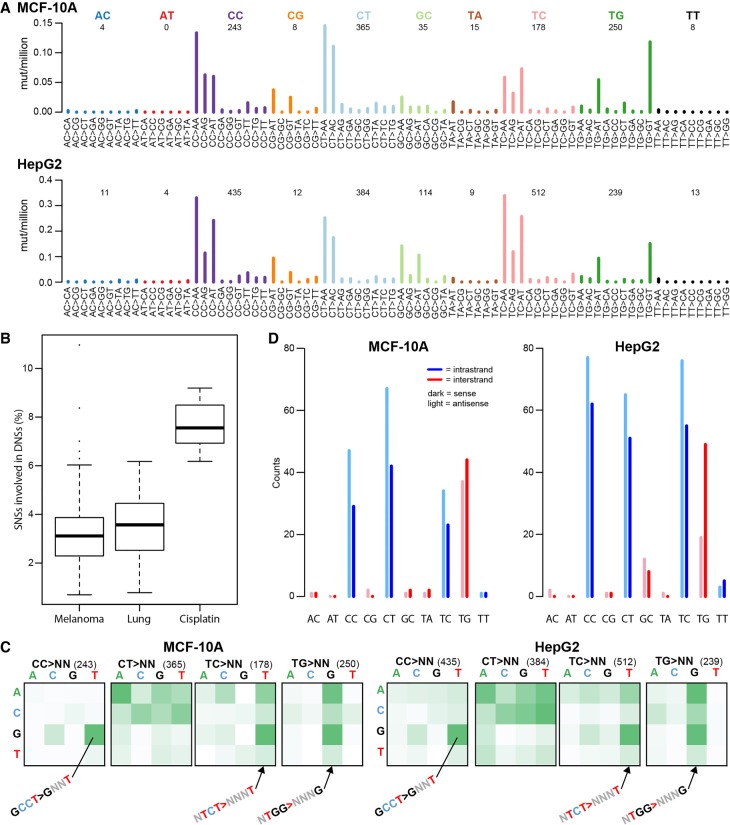
Figure 3.
Cisplatin-induced dinucleotide substitutions (DNSs). (A) DNS mutation spectra of all MCF-10A (top panel) and all HepG2 (bottom panel) clones combined, displayed as DNSs per million dinucleotides (i.e., normalized for dinucleotide abundance in the genome). (B) Cisplatin induces higher numbers of DNSs than other mutational processes associated with dinucleotide substitutions such as UV (melanoma) and smoking (lung). (C) ±1-bp sequence context preferences for the most prominent DNS mutation classes (CC > NN, CT > NN, TC > NN, and TG > NN). The total number of DNSs per mutation class is indicated in parentheses. The vertical axis is the preceding (5′) base, the horizontal axis is the following (3′) base. Some prominent enrichments in sequence context are indicated (GCCT > GNNT, NTCT > NNNT, and NTGG > NNNG). The full sequence context preference plots, both raw counts and normalized for tetranucleotide abundance in the genome, are shown in Supplemental Figure S10. (D) Transcription strand bias of dinucleotide substitutions. Potential intra-strand crosslink sites are shown in blue, potential inter-strand crosslink sites are shown in red.