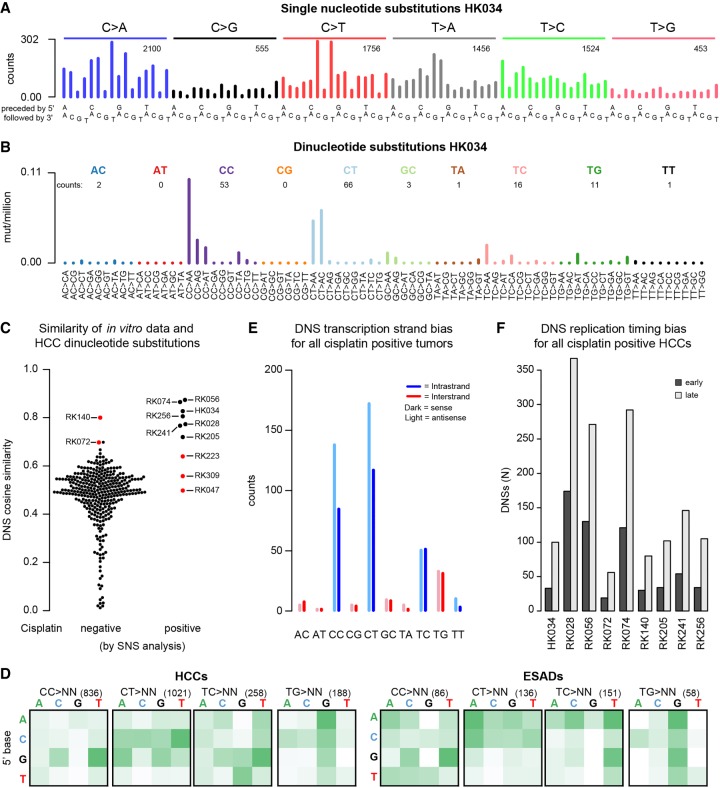
Figure 4.
Cisplatin mutational signature in human hepatocellular carcinomas (HCCs) and esophageal adenocarcinomas (ESADs). (A) Example SNS and (B) DNS mutational spectra of a tumor that tested positive for the cisplatin signature in the SNS analysis (HK034). In A and B, numbers of mutations in each mutation class are indicated. (C) DNS cosine similarities between the experimental cisplatin signature and HCCs, grouped on whether they were negative (left) or positive (right) for cisplatin mutagenesis in the SNS analysis. Red dots represent HCCs that were found positive for cisplatin mutagenesis in the SNS analysis but did not show the cisplatin DNS signature (false-positives) and samples that were not found cisplatin-positive in the SNS analysis but were concluded to be cisplatin-positive based on the DNS analysis (false-negatives). (D) ±1-bp sequence context preferences for the most prominent DNS mutation classes in cisplatin-positive HCCs and ESADs. Total numbers of DNSs per mutation class are indicated in parentheses. The vertical axis is the preceding (5′) base, the horizontal axis is the following (3′) base. (E) DNS transcription strand bias in all cisplatin-positive tumors combined. For the individual sample plots, see Supplemental Figure S27. (F) DNS replication timing bias in cisplatin-positive HCCs. DNSs were classified as being in either early or late replicating regions as described in Methods.