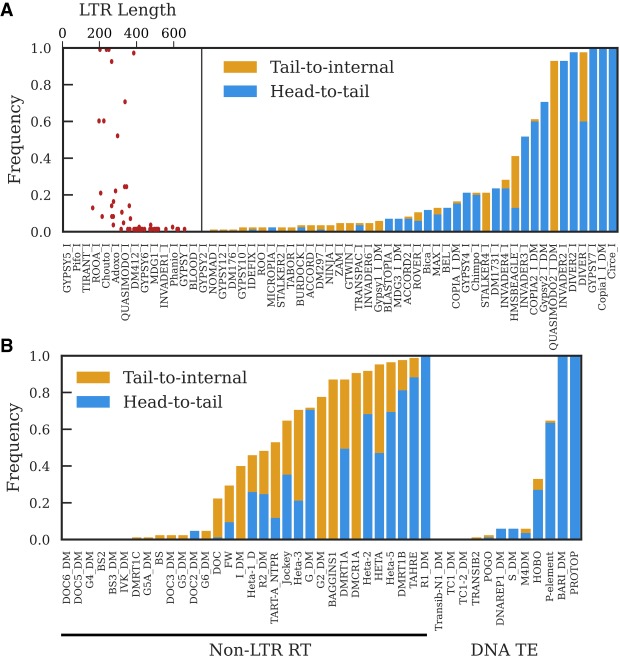
Figure 3.
The proportion of GDL strains in which a tandem junction was identified for LTR retrotransposon families (A) and non-LTR retrotransposon families and DNA transposon families (B). Head-to-tail tandems have junctions involving the first and last 200 nt of the consensus sequence. Tail-to-internal junctions have junctions between the last 200 nt of the consensus sequence and internal sequence; these are consistent with tandems involving 5′-truncated elements, though they can also be formed by nested insertions. We do not depict the frequency of internal-to-internal tandems because they are present in most strains, but generally at low copy number; Supplemental Figure S5 provides a more informative visualization of internal-to-internal tandem variation. A does not include LTR–LTR junctions shown in Supplemental Table S2. The scatter plot inset in A depicts the relationship between LTR length and the frequency of detecting head-to-tail tandems for each LTR retrotransposon family.