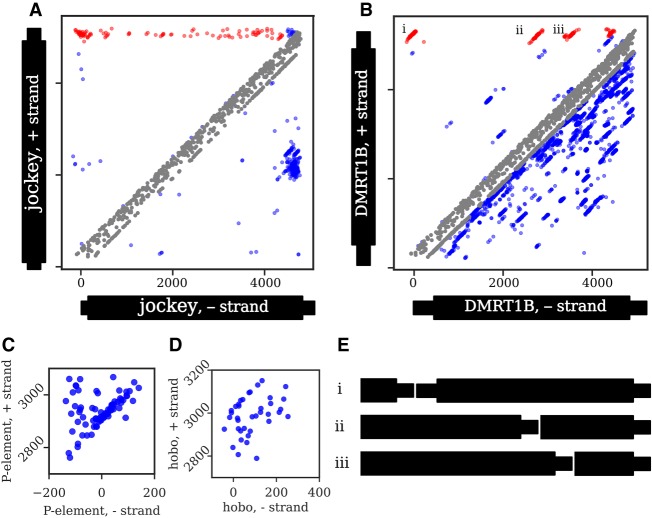
Figure 4.
Junction distributions from all strains in the GDL for two non-LTR retrotransposons (A,B) and two DNA transposons (C,D). Note that C and D only show head-to-tail tandem distributions, and thus, the axes only include the terminal regions. Each dot represents a junction identified from a single strain. A junction present in multiple strains will generate a diagonal distribution around the true coordinate due to estimation errors. In A and B, head-to-tail and tail-to-internal tandem junctions are highlighted in red, internal-to-internal tandems and deletions are colored in blue, and probable artifacts are colored in gray (see Methods, “Categorizing Tandem Junctions”); all junctions in C and D are head-to-tail. The distribution of tandem junctions of jockey (A) are dispersed, with few distinct diagonal clusters, indicating that most individual tandem junctions are low-frequency. In contrast, the four distinct diagonal clusters of DMRT1B (B) indicate junctions at moderate to high population frequency, suggesting that they represent older tandems. While not the focus of our analysis, internal deletions ranging from low to high frequency are also evident in both A and B as junctions below the main diagonal, with several distinct deletion variants of jockey sharing similar sequence coordinates and with many distinct deletions identifiable in DMRT1B. (C) For the P-element, most junctions fall within a single tight diagonal cluster, consistent with their representing tandem P-elements separated by an 8-bp target site duplication. Several junctions are dispersed above this cluster, consistent with additional sequence of variable length within the junction. (D) In contrast, only a few hobo junctions form a tight diagonal cluster, while most are dispersed below the cluster, consistent with small internal deletions spanning most of the tandem junctions. (E) Schematics of the head-to-tail and tail-to-internal DMRT1B tandems denoted with i–iii in B.