Figure 4.
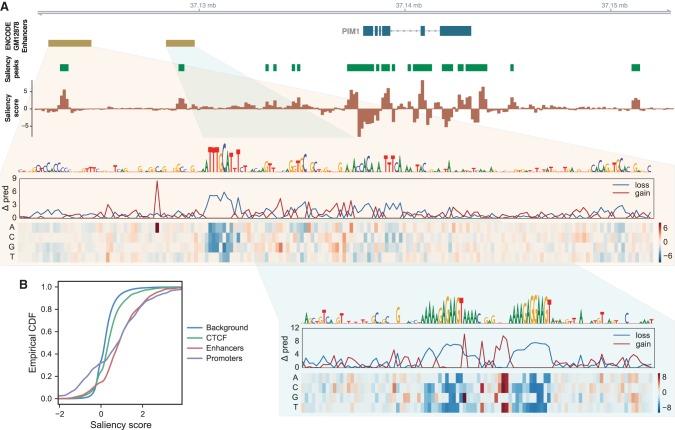
Basenji identifies distal regulatory elements. (A) ENCODE enhancer annotations for PIM1 in GM12878 specify two downstream regulatory elements. Basenji saliency scores and FDR < 0.05 peaks (see Methods) mark these elements, in addition to a variety of others that lack typical enhancer chromatin. In silico saturation mutagenesis of these elements with respect to Basenji's PIM1 GM12878 CAGE prediction outline the driving motifs. The quantities in the heatmap display the change in Basenji prediction “Δ pred” (summed across the sequence) after substituting the row's specified nucleotide into the sequence. The line plots display the minimum (loss) and maximum (gain) change among the possible substitutions. The upstream cis-regulatory module most prominently features a POU2F factor motif, while the downstream element consists solely of two adjacent PU.1 motifs. (B) We plotted the cumulative distributions of the maximum saliency score for elements of various regulatory annotation classes in GM12878 released by ENCODE. Genome-wide, each annotation class differs significantly from the background scores by Kolmogorov–Smirnov test.