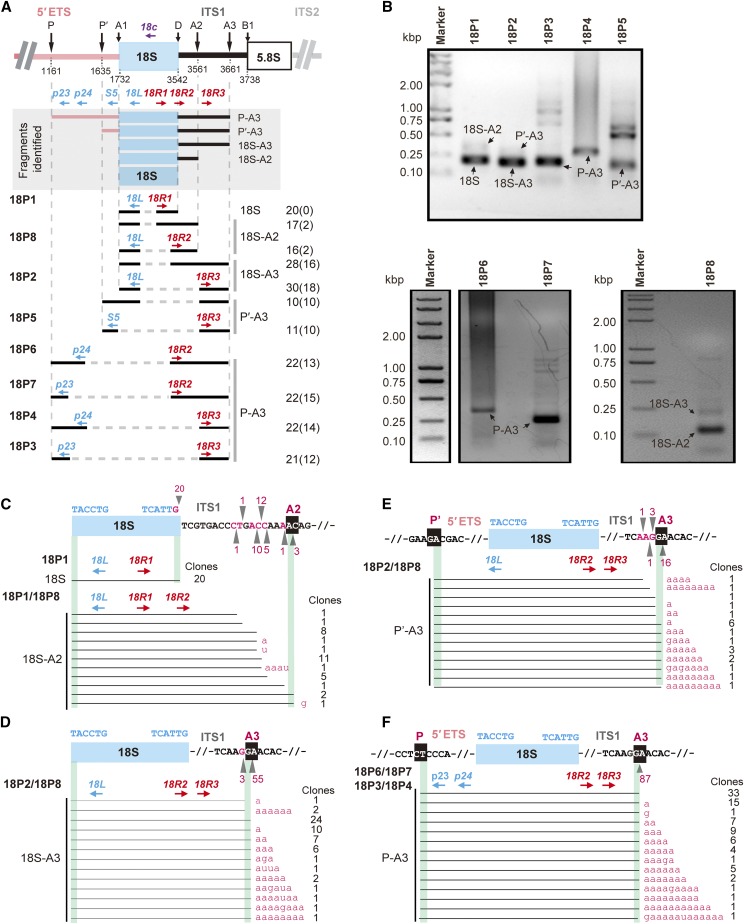
Figure 1.
Mapping of the 5′ and 3′ extremities of the pre-18S rRNAs. A, Structure of pre-18S rRNA intermediates identified by a set of primer combinations (in shaded box). Forward and reverse PCR primers for cDNA amplification are marked in red and blue, respectively. For each fragment, the number of clones obtained is indicated on the right. The number of clones with additional sequences, such as polyadenylation at the 3′ end, is marked in parentheses. Eight pairs of primers were used: 18P1 (18L/18R1), 18P2 (18L/18R3), 18P3 (p23/18R3), 18P4 (p24/18R3), 18P5 (S5/18R3), 18P6 (p24/18R2), 18P7 (p23/18R2), and 18P8 (18L/18R2). B, Pre-18S rRNA intermediates were determined in gel by cRT-PCR with primers 18P1 to 18P8. C to F, DNA sequencing of 18S and its major precursors identified: 18S-A2 (C), 18S-A3 (D), P′-A3 (E), and P-A3 (F). The 18S rRNAs identified by primers 18P1 were validated by sequencing of 20 independent clones. The 18S-A2 intermediates identified by primers 18P1 and 18P8 were validated by sequencing of 33 independent clones (C). The 18S-A3 intermediates identified by primers 18P2 and 18P8 were validated by sequencing of 58 independent clones (D). The P′-A3 intermediates identified by primers 18P2 and 18P8 were validated by sequencing of 21 independent clones (E). The P-A3 intermediates identified by primers 18P6, 18P7, 18P3, and 18P4 were validated by sequencing of 87 independent clones (F). The ITS1 locus matched by the 3′ ends of these clones are indicated by black triangles as well as the number of clones. Additional sequences in the 3′ extremities of these clones are marked in red lowercase letters. The numbers of identical clones are indicated to the right of each fragment.