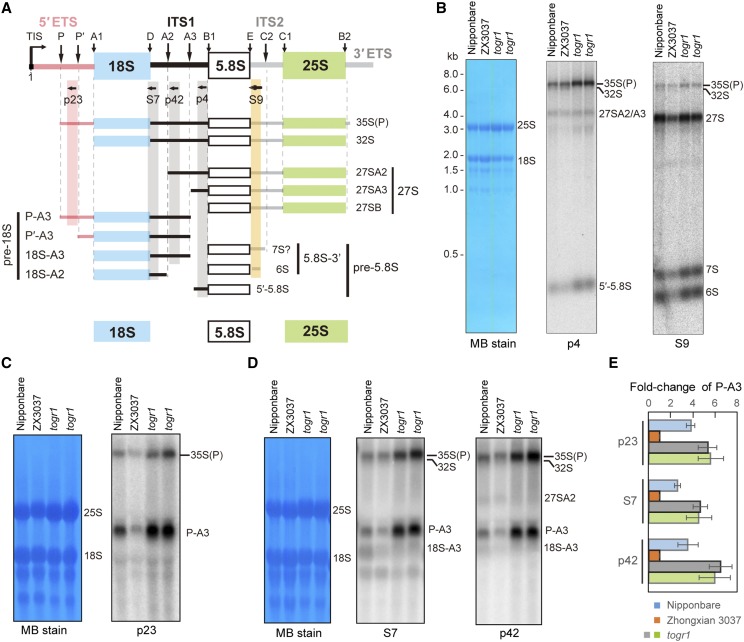
Figure 5.
Northern blots to detect pre-rRNA processing in rice. A, Pre-rRNA processing intermediates detected by northern blots with specific probes, which are indicated by horizontal arrows. Black vertical arrows above the diagram indicate endonucleolytic cleavage sites relevant to this study. Different rRNA precursors are marked. P-A3, P′-A3, 18S-A3, and 18S-A2 belong to the pre-18S rRNAs. 27SA2, 27SA3, and 27SB belong to the 27S rRNA, the common precursor of 5.8S and 25S rRNAs. The 3′-5.8S (7S and 6S) and 5′-5.8S are pre-5.8S rRNAs. The 7S rRNA marked with “?” was detected by probe S9 (Fig. 5B), but its definite 3′ extremities are still unclear (A). The 35S(P) and 27SA2 could be specifically detected by probes p23 and p42, respectively. Both probes S7 and p42 detect 35S(P), 32S, P-A3, and 18S-A3. Although 18S-A2 could be detected by S7, its low abundance in wild-type rice makes it harder to distinguish from 18S-A3 by northern-blot assay. B, Northern blots to determine pre-rRNA processing in pre-60S LSU in Nipponbare (lane 1), Zhongxian3037 (ZX3037, lane 2), and togr1 mutants (lanes 3 and 4). The togr1 mutant is a positive control that accumulates the 35S pre-rRNA and P-A3 intermediates, when compared with its wild type, Zhongxian3037 (Wang et al., 2016). Probes p4 and S9 were used. Methylene blue staining (MB stain) of the membrane is shown as the loading control. C to E, Northern blots to determine pre-rRNA processing in pre-40S SSU by probes p23 (C), S7, and p42 (D) in rice. The S7 and p42 blots share the same loading control (D). The quantitation of P-A3 in Nipponbare (lane 1), Zhongxian3037 (lane 2), and togr1 (lanes 3 and 4) were performed with three biological replicates (E). Matured rRNAs stained with MB serve as the loading control. The relative intensities for P-A3 intermediate in each lane are normalized to Zhongxian3037. Error bars represent sd. Data are given as means and sd of three independent biological replicates.