2′,3′-cAMP associates with Arabidopsis Rbp47b and plays a role in stress granule formation.
Abstract
2′,3′-cAMP is an intriguing small molecule that is conserved among different kingdoms. 2′,3′-cAMP is presumably produced during RNA degradation, with increased cellular levels observed especially under stress conditions. Previously, we observed the presence of 2′,3′-cAMP in Arabidopsis (Arabidopsis thaliana) protein complexes isolated from native lysate, suggesting that 2′,3′-cAMP has potential protein partners in plants. Here, affinity purification experiments revealed that 2′,3′-cAMP associates with the stress granule (SG) proteome. SGs are aggregates composed of protein and mRNA, which enable cells to selectively store mRNA for use in response to stress such as heat whereby translation initiation is impaired. Using size-exclusion chromatography and affinity purification analyses, we identified Rbp47b, the key component of SGs, as a potential interacting partner of 2′,3′-cAMP. Furthermore, SG formation was promoted in 2′,3′-cAMP-treated Arabidopsis seedlings, and interactions between 2′,3′-cAMP and RNA-binding domains of Rbp47b, RRM2 and RRM3, were confirmed in vitro using microscale thermophoresis. Taken together, these results (1) describe novel small-molecule regulation of SG formation, (2) provide evidence for the biological role of 2′,3′-cAMP, and (3) demonstrate an original biochemical pipeline for the identification of protein-metabolite interactors.
2′,3′-cAMP, a positional isomer of the well-known secondary messenger 3′,5′-cAMP, is an intriguing small molecule with an unknown biological function. In fact, it was only in 2009 that the existence of 2′,3′-cAMP was reported in biological material (Ren et al., 2009). Since then, 2′,3′-cAMP has been detected in both mammalian and plant cells and observed to accumulate at high levels in response to stress and injury (Jackson et al., 2009; Verrier et al., 2012; Van Damme et al., 2014). In vitro studies showed that 2′,3′-cAMP is formed during mRNA degradation, when hydrolysis of the P-O5′ bond, mediated by 3′ to 5′ RNases, is accompanied by the transphosphorylation of mRNA to form 2′,3′-cyclic nucleotides (Thompson et al., 1994). Due to the mRNA poly(A) tail, 2′,3′-cAMP reaches a high abundance in the cell. High levels of cellular 2′,3′-cAMP can have deleterious consequences (Azarashvili et al., 2009). Therefore, it is suggested that cells export 2′,3′-cAMP to the extracellular compartment, where it is metabolized to 2′-AMP and adenosine (Jackson et al., 2009; Jackson, 2017). Moreover, 2′,3′-cAMP levels correlate with adenosine content in Arabidopsis (Arabidopsis thaliana; Bernard et al., 2011). In agreement with these results, enzymes with in vitro 2′,3′-cyclic nucleotide 3′-phosphodiesterase activity were reported in both animals (Vogel and Thompson, 1987) and plants (Culver et al., 1994; Genschik et al., 1997).
Considering the limited understanding of 2′,3′-cyclic nucleotide metabolism and function, our recent findings, namely the demonstration of 2′,3′-cAMP and 2′,3′-cGMP presence in protein complexes isolated from native Arabidopsis lysate, are indeed of great interest (Veyel et al., 2017). Using size-exclusion chromatography (SEC) to separate protein-bound metabolites from free metabolites, we showed that both 2′,3′-cAMP and 2′,3′-cGMP coelute with proteins of approximately 40 kD. Intrigued by this observation, we decided to uncover the identity of the protein interactor(s), focusing first on the more abundant 2′,3′-cAMP, with the assumption that its interactors might be shared with 2′,3′-cGMP. We identified Rbp47b (encoded by At3g19130) as the most promising candidate for a 2′,3′-cAMP receptor. Rbp47b is a polyadenylate-binding protein known to be involved in RNA binding and aggregation in the cytoplasm, participating in the formation of stress granules (SGs) under stress conditions (Kedersha et al., 1999; Lorković et al., 2000; Weber et al., 2008). SGs are large aggregates of proteins and untranslated mRNAs first reported in heat-treated tomato (Solanum lycopersicum) cells (Nover et al., 1989; Buchan and Parker, 2009). The main role of SGs is to establish and subsequently moderate the stress translatome by selective storage and protection of the mRNAs. This role was hypothesized by Nover et al. (1989) and proven subsequently by numerous studies (Yan et al., 2014; Khong et al., 2017).
Due to their key importance in the cellular stress response, SG assembly and disassembly have been studied intensively (Anderson and Kedersha, 2008; Wheeler et al., 2016). One of the events crucial for SG assembly is phosphorylation of the translation initiation factor eIF2, which impedes the association of tRNAMet to the 43S preinitiation complex, leading to translational repression. Rather than undergoing translation, mRNA molecules of the 43S complex associate with RNA-binding proteins, such as Rbp47b, which supports protein aggregation and SG formation (Kedersha et al., 1999; Weber et al., 2008). A comprehensive protein composition of SGs was published recently for yeast and mammalian cells (Jain et al., 2016), revealing a stable core composed of 40S ribosomal subunits, translation initiation factors, RNA-binding proteins (RBPs), chaperones (heat shock proteins and CCT complex), metabolic enzymes, RNA/DNA-remodeling complexes, and signaling proteins.
SG formation is tightly controlled. To date, the described regulatory mechanisms include (1) posttranslational modifications of key SG components (Arimoto-Matsuzaki et al., 2016), (2) ATP- and ATP-driven remodeling complexes (Jain et al., 2016), and (3) microtubule-dependent regulation of SG movement and disassembly (Gutierrez-Beltran et al., 2015). Here, we present a novel putative SG regulatory mechanism that involves a small-molecule ligand, namely 2′,3′-cAMP, which binds to Rbp47b and facilitates SG formation.
RESULTS
SG-Associated Proteins Are Overrepresented among Putative 2′,3′-cAMP Interactors
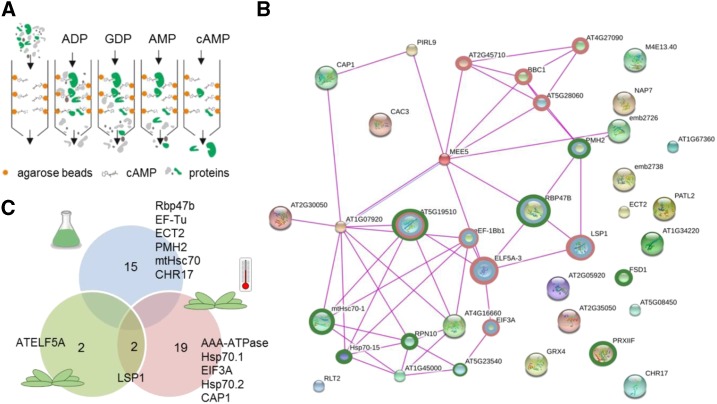
To identify binding protein partner(s) of 2′,3′-cAMP, we adapted a previously described affinity purification (AP) protocol (Aye et al., 2009) and used agarose beads coupled to 2′,3′-cAMP via the NH2 group of the purine ring (Supplemental Fig. S1). After incubation of the beads with total soluble-protein lysate (referred to as input), unspecific binding proteins were removed by sequential washes with ADP, GDP, and 5′-AMP, followed by specific elution with 2′,3′-cAMP (Fig. 1A). Both washes and eluate were analyzed by liquid chromatography-mass spectrometry (LC-MS/MS) proteomics.
Figure 1.
AP analysis using 2′,3′-cAMP agarose beads showed enrichment in SG proteins. A, Schematic representation of the AP analysis. B, Schematic representation of putative 2′,3′-cAMP interactors retrieved from the Stitch database (Szklarczyk et al., 2016). Experimental evidence and a low-confidence cutoff were used to visualize protein-protein interactions. Proteins involved in translation (red rings), RNA binding (blue rings), and stress response (green rings) are highlighted. C, Venn diagram containing proteins identified as putative 2′,3′-cAMP binders in cell cultures (blue), Arabidopsis rosette leaves (green), and rosettes under heat stress (red). Proteins putatively annotated as SG components based on the human SG composition are indicated (Jain et al., 2016). The data presented are from three (cell cultures) or four (rosettes) technical replicates separated at the lysate stage.
Following our previously published (Veyel et al., 2017) results demonstrating that 2′,3′-cAMP is retained in protein complexes isolated from Arabidopsis cell cultures, we used MM2d cells as an initial source of starting material. Proteins more abundant in the eluate in comparison with the washes (Student’s t test, P < 0.1) were defined as 2′,3′-cAMP-binding proteins (Fig. 1B; Supplemental Table S1). A total of 15 proteins passed the cutoff, seven of which eluted from the agarose beads exclusively with 2′,3′-cAMP but were absent in any of the washes, including the polyadenylate-binding protein Rbp47b (Fig. 1C; Supplemental Table S1), a key component of SGs (Kedersha et al., 1999; Weber et al., 2008).
To broaden our search for 2′,3′-cAMP-interacting proteins, an additional AP analysis was performed using 5-week-old Arabidopsis rosettes either grown under control conditions or exposed to 30 min of heat stress (42°C), the latter of which is known to elevate 2′,3′-cAMP levels (discussed in the next section). Using the criteria mentioned above, four and 21 proteins were identified as 2′,3′-cAMP-binding interactors, respectively (Fig. 1B; Supplemental Table S2). Two proteins were commonly detected in the rosette samples, but no overlap was observed with the cell culture experiment (Fig. 1C).
To analyze the possibility of shared functionality, we plotted a total of 38 putative 2′,3′-cAMP-binding proteins identified in the three AP analyses using the Stitch database (Szklarczyk et al., 2016; Fig. 1C). Proteins belonging to the categories RNA binding, translation, and response to stress were well represented. These comprised 40S and 60S ribosomal subunits, translation initiation factors, heat shock proteins, the RNA helicase PMH2, and the RNA-binding protein Rbp47b. When we used the three functional categories as keywords, we retrieved a common denominator: SGs. Whereas Rbp47b is a known component of plant SGs (Kedersha et al., 1999; Weber et al., 2008), 12 additional putative 2′,3′-cAMP interactors have homologs found in the human SG proteome (Li et al., 2010; Jain et al., 2016; Supplemental Table S3). These are translation initiation factors, an AAA-type ATPase, heat stress proteins, a cyclase-associated protein (CAP1), an RNA helicase (PMH2), and a chromatin remodeling factor (CHR17). To evaluate the specificity of the identified 2′,3′-cAMP interactors, we queried our data against a list of 119 putative 3′,5′-cAMP and 3′,5′-cGMP binders identified in an analogous set of AP analyses (Donaldson et al., 2016). Only a single ribosomal protein (At2g45710) was found in the overlap, thus illustrating the specificity of the 2′,3′-cAMP interactor list.
In humans, the SG proteome was demarcated using transgenic lines expressing GFP-G3BP protein. A series of differential centrifugations followed by AP using anti-GFP antibody was performed in control and sodium arsenite-treated cells, a stress condition known to promote SG formation (Jain et al., 2016). Approximately 300 proteins that were at least twice more abundant in the stressed cells than in the control cells were defined as putative SG components. Interestingly, when we applied a similar cutoff to the one employed by Jain et al. (2016) and compared the proteins identified in the 2′,3′-cAMP eluate with heat-treated versus control rosettes, we found a set of 85 proteins with increased abundance (Supplemental Table S4). Of these, three were reported previously in Arabidopsis to be components of SG cores, namely Rbp47b (Weber et al., 2008), PAB8 (Sorenson and Bailey-Serres, 2014), and TUDOR-SN2 (Gutierrez-Beltran et al., 2015), and 23 more have homologs found in the human SG proteome (Lee et al., 2014; Jain et al., 2016).
In summary, 2′,3′-cAMP AP analyses identified a total of 38 putative 2′,3′-cAMP interactors. Functional analysis suggested a connection between 2′,3′-cAMP and SG assembly.
2′,3′-cAMP Accumulation Coincides with and Promotes SG Formation
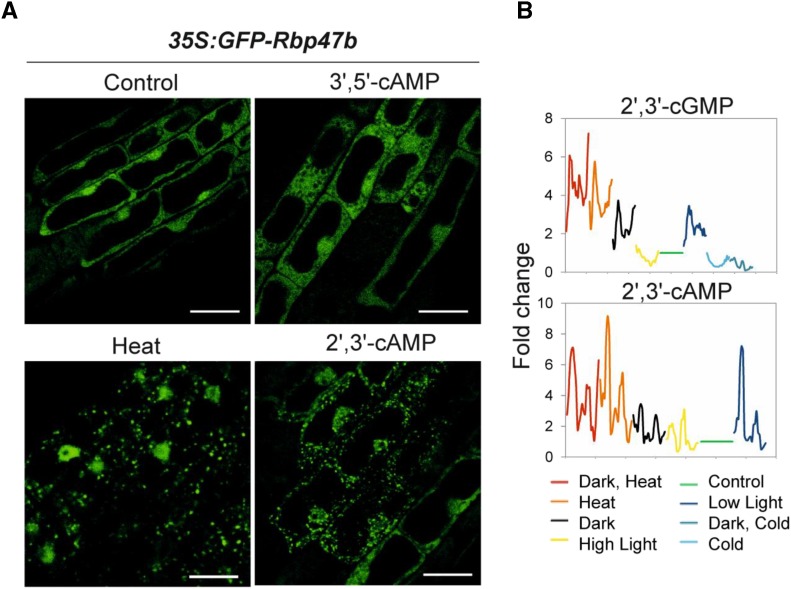
Our AP analysis revealed a connection between 2′,3′-cAMP and SGs. To functionally examine this association, we treated the Arabidopsis SG marker line 35S:GFP-Rbp47b (Gutierrez-Beltran et al., 2015) with the membrane-permeable 2′,3′-cAMP analog Br-2′,3′-cAMP. Following this, confocal microscopy was used to examine GFP-Rbp47b localization in Arabidopsis root cells. Thirty minutes to 1 h of treatment with 100 μm Br-2′,3′-cAMP led to the formation of SGs in 25% of the examined seedlings, whereas no effect was observed in either control samples (mock treated) or plants treated with 100 μm Br-3′,5′-cAMP (the closest homolog of Br-2′,3′-cAMP; Fig. 2A; Supplemental Fig. S2; Supplemental Table S5). Heat treatment was used as a positive control and, as expected, all the cells contained SGs.
Figure 2.
2′,3′-cAMP accumulates in response to stress conditions and promotes SG formation. A, Subcellular localization of the GFP-Rbp47b protein under different experimental conditions (after 1 h of treatment). Additional micrographs are given in Supplemental Figure S2. Bars = 10 μm. B, 2′,3′-cGMP and 2′,3′-cAMP accumulation in mature Arabidopsis rosettes under different stress conditions. Six-week-old Arabidopsis plants were shifted to either control or stress conditions. First, samples (whole rosette) were taken at 5, 10, and 20 min following stress onset, and afterward, every 20 min until 6 h. The last two samples were taken at 10 and 20 h after stress onset. Graphs represent the accumulation of 2′,3′-cGMP and 2′,3′-cAMP over time in different stresses (color legend at bottom). Data are presented as fold change (rolling average) calculated in comparison with the paired control. Each data point is the average of three biological replicates (individual rosettes). Control temperature and light conditions (21°C and 150 µE m‒2 s‒1) were used as a paired control for heat stress (32°C), low light (70 µE m‒2 s‒1), high light (300 µE m‒2 s‒1), cold (4°C and 85 µE), and darkness treatments. Note that 2′,3′-cAMP was not detected under cold stress conditions.
As an additional line of evidence, we mined metabolomics data from a previously published time-course experiment (Caldana et al., 2011) and analyzed whether 2′,3′-cAMP levels change in stress conditions known to promote SG formation. Briefly, mature Columbia-0 (Col-0) rosettes (prior to bolting) grown in a controlled environment were transferred into combinations of light (low light, high light, and darkness) and temperature (heat and cold) conditions. Samples were collected at multiple time points until 24 h following experimental treatment onset. Analysis of secondary metabolites revealed that 2′,3′-cAMP but also 2′,3′-cGMP accumulated in low light and darkness, most notably under heat stress (Fig. 2B), which are all conditions known to promote SG assembly (Gutierrez-Beltran et al., 2015). Moreover, the timeline of 2′,3′-cyclic nucleotide accumulation showed a rapid increase measured in the first 5 to 10 min of the treatment, coinciding well with the previously reported timing of SG assembly of 10 to 20 min after stress onset (Gutierrez-Beltran et al., 2015).
In summary, our results support a functional connection between 2′,3′-cAMP and SG assembly. Increased 2′,3′-cAMP levels coincide with (stress time-course experiment) and promote (2′,3′-cAMP treatment experiment) SG formation.
2′,3′-cAMP and Rbp47b Cofractionate in SEC Analysis
Three AP analyses revealed a total of 38 putative direct/indirect 2′,3′-cAMP interactors. To narrow down this list, we decided to integrate it with SEC data that suggested that 2′,3′-cAMP is retained in protein complexes in size fractionation experiments (Veyel et al., 2017).
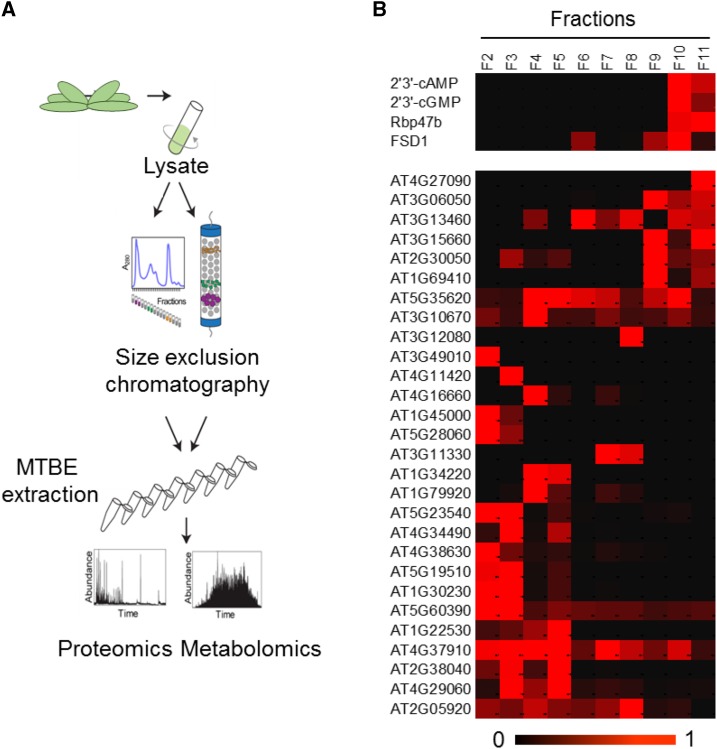
To find a direct 2′,3′-cAMP protein partner, native soluble lysate was prepared from Arabidopsis rosettes (Fig. 3A). Protein-protein and protein-metabolite complexes of approximately 700 to 20 kD were separated using SEC. Obtained fractions were extracted using the methyl tert-butyl ether (MTBE) method (Giavalisco et al., 2011) and analyzed with respect to their metabolite and protein composition to identify possible interactors. The quality of our experimental data set was confirmed by analyzing the coelution profile of proteasome subunits, which served as a control in a previously reported study (Aryal et al., 2014). From a total of 20 identified subunits, 18 cofractionated together with the maximum intensity measured in the two neighboring fractions (F2 and F3), corresponding to protein macrocomplexes of approximately 700 kD (Supplemental Fig. S3).
Figure 3.
Rbp47b coelutes with 2′,3′-cAMP in SEC analysis. A, Schematic representation of the SEC analysis. B, Heat map representing elution profiles of the 2′,3′-cyclic nucleotides across protein-containing fractions normalized to the maximum intensity. The elution behavior of proteins identified as 2′,3′-cAMP putative interactors in the AP analysis is shown. Rbp47b and FSD1 proteins clearly coelute with the 2′,3′-cyclic nucleotides. For the color scale bar at bottom, 1 = present and 0 = absent. The data presented represent three (metabolite) or one (protein) biological replicates.
In line with our previously published study (Veyel et al., 2017), 2′,3′-cAMP and 2′,3′-cGMP were reproducibly present in protein-containing fractions (F10 and F11) that correspond to 50- to 20-kD proteins (Fig. 3B). To find proteins with an elution profile similar to that of 2′,3′-cyclic nucleotides, we applied a Pearson correlation coefficient cutoff higher than 0.7. In addition, protein presence in the F10 and F11 fractions also was required. Analysis identified 191 potential 2′,3′-cyclic nucleotide interactors. We proposed that at least one represented a true binder (Supplemental Table S6).
Comparison with the AP list revealed an overlap comprising two proteins (Fig. 3B): the RNA-binding protein Rbp47b and an iron superoxide dismutase (FSD1). Both proteins were enriched in the cell-culture AP analysis. In fact, Rbp47b (but not FSD1) also was reproducibly present in the 2′,3′-cAMP eluate from the rosette experiments, although its abundance was not significantly higher than in the washes (Supplemental Fig. S4). These results strongly support Rbp47b, a key component of Arabidopsis SGs, as the most promising 2′,3′-cAMP interactor candidate.
2′,3′-cAMP Binds to Rbp47b via RRM2 and RRM3 Domains
Although the results of both AP and SEC analyses clearly indicated Rbp47b as a major 2′,3′-cAMP partner, confirmation of direct in vitro binding was necessary. For that purpose, Rbp47b was cloned, expressed, and purified in Escherichia coli (Supplemental Fig. S5A). Protein folding was examined using nanoDSF technology (Supplemental Fig. S5B), which relies on tracing protein thermal stability by recording changes in Trp residues in an increasing temperature gradient. Based on the shape of the melting curve, we could conclude that the protein is properly folded, and the melting temperature (Tm) of Rbp47b was calculated at 55.6°C (Supplemental Figs. S5B and S6). As Tm changes in the presence of a small-molecule ligand are indicative of binding, we tested the effect of 2′,3′-cAMP and 2′,3′-cGMP on the thermal stability of Rbp47b. At 2 mm, 2′,3′-cAMP but not 2′,3′-cGMP did, in fact, stabilize Rbp47b by 0.3°C (Supplemental Fig. S6B). Protein purity was confirmed through mass spectrometry analysis (Supplemental Fig. S5C).
Although nanoDSF attests to binding between Rbp47b and 2′,3′-cAMP (small but significant shift in Tm), no definite conclusion could be drawn regarding the affinity and specificity of the interaction. Therefore, we attempted to determine the in vitro binding affinity using microscale thermophoresis (MST; Jerabek-Willemsen et al., 2011). MST exploits the effect of size, charge, and hydration shell on the movement of molecules in a temperature gradient. Complex formation resulting in the alteration of at least one of the three parameters leads to differential movement, from which the binding affinity can be determined.
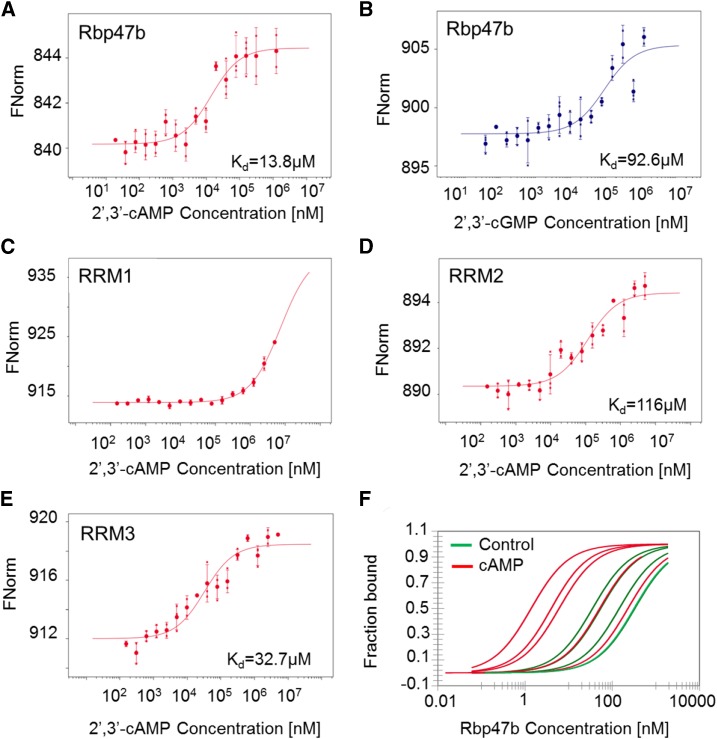
The MST results confirmed 2′,3′-cAMP binding to Rbp47b (Fig. 4A), with a Kd value of 14 µm. Most importantly, we did not observe any binding for 3′,5′-cAMP, indicating very high specificity of Rb47b for 2′,3′-cAMP (Supplemental Fig. S7; for traces, see Supplemental Fig. S8). In addition, 2′,3′-cGMP, but not 3′,5′-cGMP, bound Rbp47b (Fig. 4B; Supplemental Fig. S7). However, the 93 µm Kd of 2′,3′-cGMP binding was significantly higher than the Kd measured for 2′,3′-cAMP, indicating the preference of Rbp47b for 2′,3′-cAMP. The measured Kd value also corresponds to the low-to-mid micromolar concentration of 2′,3′-cAMP measured in Arabidopsis lysate (Supplemental Fig. S9).
Figure 4.
In vitro validation of the Rbp47b-2′,3′-cAMP interaction. A, MST measurements of the interaction between Rbp47b and 2′,3′-cAMP. B, MST measurements testing the interaction between Rbp47b and 2′,3′-cGMP. C, MST measurements testing the interaction between RRM1 and 2′,3′-cAMP. D, MST measurements testing the interaction between RRM2 and 2′,3′-cAMP. E, MST measurements testing the interaction between RRM3 and 2′,3′-cAMP. F, MST measurements of Rbp47b self-assembly in the presence of 500 μm 2′,3′-cAMP. Data are presented as bound and nonbound proteins. Data represent independent titrations. For A to E, data are averages ± sd, n = 3 (data are from independent titrations). FNorm corresponds to normalized fluorescence of labeled Rbp47b.
Rbp47b is composed of three RRM RNA-binding motifs, known to recruit mRNAs (Supplemental Fig. S10), and the prion-like PRD domain that supports protein-protein interactions (Gilks et al., 2004; Weber et al., 2008). Interestingly, the RRM domain shares topological similarity with the RNA-binding domain of the putative 2′,3′-cAMP phosphodiesterase (Hofmann et al., 2000). Thus, we speculated that 2′,3′-cAMP will bind to one or more Rbp47b RRM motifs.
The three RRM domains RRM1, RRM2, and RRM3 were expressed and purified in E. coli (Supplemental Fig. S11). As done previously, proper folding of all the domains was examined using nanoDSF, and 2′,3′-cAMP binding was assayed using MST. Whereas no binding was measured for RRM1, both RRM2 and RRM3 bound 2′,3′-cAMP, with Kd values of 116 and 33 µm, respectively (Fig. 4, C‒E).
Finally, we tested whether 2′,3′-cAMP would have an effect on Rbp47b self-oligomerization, a key event in SG formation (Gutierrez-Beltran et al., 2015). Using unlabeled Rbp47b as a ligand, we performed MST measurements (Fig. 4F). The obtained Kd of the self-interaction varied from 328 to 35 nm (median from five independent titrations, 144 nm). Moreover, the addition of saturating amounts of 2′,3′-cAMP shifted the Kd to lower values, but once more the Kd of the self-interaction varied considerably from 228 to 2 nm (median from five independent titrations, 6 nm).
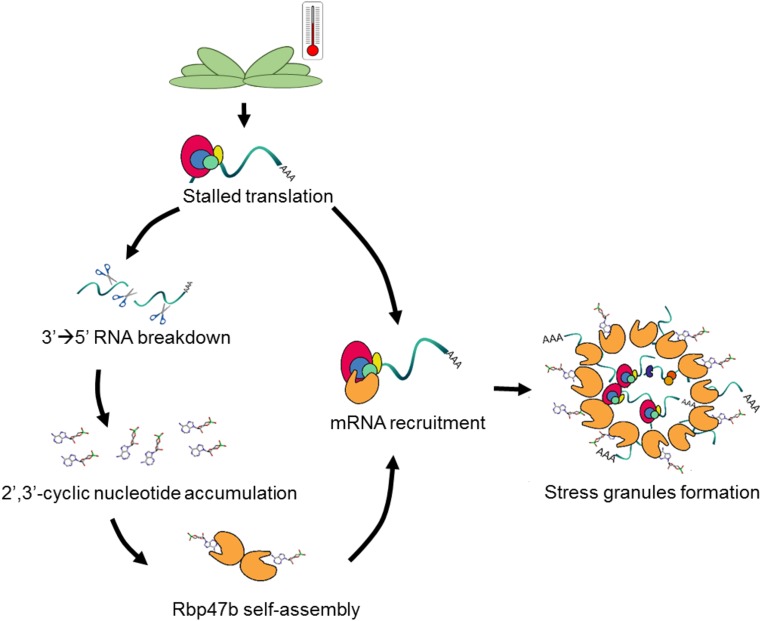
Taken together, we propose a model (Fig. 5) in which Rbp47b protein binds to 2′,3′-cyclic nucleotides (but not to 3′,5′-cyclic nucleotides) under stress conditions due to its accumulation in the cytosol. Two domains of Rbp47b, namely RRM2 and RRM3, constitute a 2′,3′-cAMP-binding site. As a consequence of binding, 2′,3′-cAMP lowers the Kd of Rbp47b self-assembly, facilitating SG formation.
Figure 5.
Proposed mechanism of 2′,3′-cAMP in SG formation due to its interaction with Rbp47b. Stress conditions (e.g. heat) induce translational arrest as well as RNA breakdown. The activity of the 3′ to 5′ RNA nucleases leads to the accumulation of 2′,3′-cyclic nucleotides, which consequently bind to Rbp47b. The binding of 2′,3′-cAMP and mRNA facilitates Rbp47b self-assembly, thereby promoting SG formation.
DISCUSSION
2′,3′-cAMP: A Novel Regulator of SG Assembly
The first main finding of our study is the association between 2′,3′-cAMP and SG formation. Initial evidence was obtained in AP analyses (2′,3′-cAMP agarose beads; Fig. 1), where an enrichment in SG core-associated proteins (40S ribosomal subunits, translation initiation factors, heat shock proteins, and RNA-binding proteins) was observed (Jain et al., 2016). This was particularly noticeable when we compared 2′,3′-cAMP eluate from control and heat-treated rosettes, heat being a well-known trigger for SG formation (Weber et al., 2008).
We could demonstrate that, under heat and dark stress conditions, plants accumulate cellular 2′,3′-cAMP and 2′,3′-cGMP. Rapid increases of 2′,3′-cAMP and 2′,3′-cGMP levels upon the mentioned stresses coincide with the timing of SG formation (Fig. 2). Since co-occurrence on its own does not prove causation, we evaluated whether 2′,3′-cAMP might trigger SG assembly in vivo (Fig. 2A). Our results revealed that 2′,3′-cAMP treatment was sufficient to promote SG formation in Arabidopsis root cells. Intriguingly, although the induction of SGs by 2′,3′-cAMP was clear, in contrast to heat stress, not all of the examined seedlings responded equally to the treatment. This observation can be explained by possible differential diffusion of 2′,3′-cAMP across cellular membranes. Furthermore, it is likely that 2′,3′-cAMP might trigger SG formation only if the cells are in a 2′,3’-cAMP-competent state, meaning that possibly a subthreshold stress level is required. In such a scenario, 2′,3′-cAMP would not be a primary trigger for SG formation but rather a facilitator of SG assembly.
The biological role of 2′,3′-cAMP is poorly characterized, particularly in plants. In animals, 2′,3′-cAMP is discussed as a toxic by-product of mRNA degradation, rapidly converted to 2′-AMP and adenosine (Jackson, 2017). Mice defective in the activity of 2′,3′-cyclic nucleotide phosphodiesterase (CNPase), a marker enzyme of myelin and oligodendrocytes, are characterized by a number of histological and functional brain abnormalities, leading to faster aging and the onset of neurodegenerative diseases (Lappe-Siefke et al., 2003). This is, at least to some extent, related to the ability of 2′,3′-cAMP to accelerate the opening of mitochondrial transition pores and, as a consequence, trigger apoptosis (Azarashvili et al., 2009). Interestingly, SG accumulation, which at least in Arabidopsis is facilitated by 2′,3′-cAMP, also was linked to both neurodegeneration and aging (Lechler and David, 2017), possibly providing an additional explanation for the phenotype observed in the CNPase knockout mice.
In light of our results, we decided to characterize the interaction between 2′,3′-cAMP and Rbp47b (Fig. 3). Using biophysical approaches, we observed the specificity and high-affinity binding of 2′,3′-cAMP to Arabidopsis Rbp47b (Fig. 4, A and B). A Kd value of 14 µm, measured for the Rbp47b-2′,3′-cAMP interaction, is within the binding-affinity range reported for various receptor-ligand interactions, such as between the GA receptor GID1 and 16,17-dihydro-GA4 (Kd = 1.4 µm) and between abscisic acid and the PYR1 receptor (Kd = 97 µm; Ueguchi-Tanaka et al., 2005; Dupeux et al., 2011). Interestingly, the measured Kd also is in agreement with the cellular 2′,3′-cAMP concentration (low-to-mid micromolar range), suggesting that, indeed, this interaction might occur in the cell.
Following the interaction study, the next step to understand the biological function of Rbp47b was to identify the domain responsible for the binding of 2′,3′-cAMP. Rbp47b is composed of three RRM RNA-binding motifs, known to recruit mRNAs, and of a prion-like PRD domain that supports protein-protein interactions (Gilks et al., 2004; Weber et al., 2008). Based on sequence similarity with the putative 2′,3′-cAMP phosphodiesterase (Hofmann et al., 2000), we anticipated 2′,3′-cAMP binding to one of the Rbp47b RRM motifs (Fig. 4, C‒E). In fact, two of the RRM domains, namely RRM2 and RRM3, can bind 2′,3′-cAMP in vitro. Interestingly, binding affinities measured for RRM2 and RRM3 were lower than that measured for the full-length protein, suggesting a synergistic binding behavior. Notably, our results are reminiscent of the RNA-binding properties reported for Rbp47b (Lorković et al., 2000). Although both RRM2 and RRM3 (but not RRM1) can interact with RNA stretches, the RRM2-RRM3 tandem has a much higher affinity to RNA than single domains.
RRMs and PRD are equally important for SG formation, as demonstrated by domain-deletion studies (Gilks et al., 2004; Weber et al., 2008). While RRMs bind nontranslating RNAs, the PRD domain mediates Rbp47b self-assembly and, thus, aggregate formation (Gilks et al., 2004; Weber et al., 2008). Importantly, the PRD domain is not sufficient to form aggregates in vivo, pointing to the importance of RNA binding for SG assembly. We hypothesize that 2′,3′-cAMP binding to RRM2 and RRM3 could have a similar function and facilitate the aggregation process by decreasing the Kd of Rbp47b self-assembly (Fig. 4F).
But why would a cell need a small molecule to compete with RNA binding and mimic its action? For the moment, we can only speculate that this can be traced to the origin of 2′,3′-cyclic nucleotides. As products of RNA turnover, and in parallel facilitators of SG assembly, 2′,3′-cyclic nucleotides may help to regulate the balance between RNA degradation and storage. In fact, the same stress conditions that trigger 2′,3′-cyclic nucleotide accumulation and SG formation also are known to increase RNA degradation rates (Baginsky and Gruissem, 2002; Merret et al., 2013).
Finally, an open question remains whether Rbp47b is the sole 2′,3′-cAMP interactor. We speculate that this is rather unlikely. Rbp47b, together with other SG-related proteins, such as the poly(A)-binding proteins PAB2 and PAB8 and the oligouridylate-binding proteins UBP1A to UBP1C, belongs to a large RNA-binding protein family characterized by the presence of well-conserved RRM2-RRM3 motifs (Chantarachot and Bailey-Serres, 2018). Indeed, in our AP rosette analysis, PAB2 and PAB8 were reproducibly found in the 2′,3′-cAMP eluate, whereas Rbp45b and Rbp45c cofractionated with 2′,3′-cyclic nucleotides in the SEC analysis. Moreover, whereas Rbp47b shows higher binding affinity toward 2′,3′-cAMP, other speculated binders may have preference for the remaining 2′,3′-cyclic nucleotides, adding to the complexity of the 2′,3′-cyclic nucleotide protein interactome.
In summary, our work assigns a novel biological role to the 2′,3′-cAMP small molecule. At least in Arabidopsis, 2′,3′-cAMP facilitates SG formation, a role likely to be evolutionarily conserved across different organisms.
MATERIALS AND METHODS
Arabidopsis Cell Cultures and Plant Growth
Arabidopsis (Arabidopsis thaliana) cells cultures (Menges and Murray, 2002) were grown in MSMO medium supplemented with 3% [v/v] Suc, 0.05 mg L−1 kinetin, and 0.5 mg L−1 1-naphthaleneacetic acid on an orbital shaker at 130 rpm in the light. Cells were passaged weekly to fresh medium and harvested during logarithmic growth using rapid filtration and liquid nitrogen snap freezing. Prior to the AP and SEC analyses, Arabidopsis Col-0 was grown in soil for 4 weeks in short days (8 h, 120 µE light intensity) and then transferred for 1 week to long-day conditions (16 h, 150 µE light intensity) at day/night temperatures of 20°C/18°C. After this time, Col-0 rosettes were harvested, immediately frozen using liquid nitrogen, and pulverized using a Retsch mill for three periods of 1 min each at 60 rps. In addition, for AP, Arabidopsis plants were subjected to 30 min of control or heat stress treatment (42°C).
Preparation of Native Arabidopsis Lysate for the AP Experiment
Plant cell material was collected on the filter and pulverized to homogeneity in liquid nitrogen with mortar and pestle. In the case of rosettes, leaves from 20 Arabidopsis plants were harvested and frozen in liquid nitrogen. Homogenization was followed by resuspension in 1 mL of lysis buffer (25 mm Tris-HCl, pH 7.5, 0.5 m NaCl, 15 mm MgCl2, 0.5 mm DTT, 1 mm NaF, 1 mm Na3VO4, and 1× Protease Inhibitor Cocktail [Sigma-Aldrich; P9599]) per 1 g of plant material per sample (three to four technical replicates). Cellular debris was separated by 10 min of centrifugation at 4°C and 4,000g. Crude lysate was subjected to ultracentrifugation (45 min, 4°C, and 137,200g) to obtain a soluble fraction referred to as the native Arabidopsis lysate. Replicates were separated at the lysate stage before performing the AP experiment.
AP
Custom 2′,3′-cAMP agarose beads were purchased from Cube Biotech. 2′,3′-cAMP was coupled to the beads using the amine (NH2) group of the purine ring and a 14-carbon spacer arm. Before use, beads were equilibrated with lysis buffer. A total of 3 mL of native lysate from the cell culture (approximately 75 mg of total protein) and rosettes (approximately 30 mg) was combined with 150 µL of agarose resin (see above), incubated for 1 h on a rotating wheel at 4°C (binding), transferred to a Mobicol Classic (35-µm pore size filter) column, and washed with 10 mL of wash buffer (0.025 m Tris-HCl, pH 7.5, and 0.5 m NaCl). A total of 400 µL of 1 mm ADP (Sigma-Aldrich; 01905) dissolved in lysis buffer was added to the beads, followed by 1 h of incubation at 4°C in a table shaker (112g). Eluate was collected, and beads were washed with 10 mL of wash buffer. The procedure was repeated using 1 mm GDP (Sigma-Aldrich; G7127), 1 mm 5′-AMP (Sigma-Aldrich; A2252), and 1 mm 2′,3′-cAMP (Sigma-Aldrich; A9376). Proteins were precipitated using 2.5 volumes of prechilled acetone. Protein pellets were dried in a vacuum concentrator and stored at −20°C.
Preparation of Native Arabidopsis Lysate for SEC
Col-0 Arabidopsis rosettes were harvested from three separate trays constituting three biological replicates. The cell lysis and extraction protocol was adjusted from Veyel et al. (2017). A total of 0.7 mL of lysis buffer (50 mm AmBIC, 150 mm NaCl, 1.5 mm MgCl2, 5 mm DTT, 1 mm PMSF, 1× Protease Inhibitor Cocktail [Sigma-Aldrich], 0.1 mm Na3VO4, and 1 mm NaF) was added per 1 g of plant material. The mixture was gently vortexed (with two metal balls) until thawed and centrifuged for 20 min at 4,000g and 4°C (Beckman Coulter). The obtained supernatant was transferred to ultracentrifuge tubes and centrifuged for 1 h at 35,000 rpm (maximum, 148,862g; average, 116,140g) and 4°C, resulting in a soluble fraction containing endogenous complexes. The soluble fraction was loaded on an Amicon Ultra-15 Centrifugal filter unit (10-kD molecular mass cutoff) prerinsed with 15 mL of wash buffer (50 mm AmBIC, 150 mm NaCl, and 1.5 mm MgCl2) and centrifuged for 20 min at 4,000g and 4°C. The experiment was performed in triplicate. Metabolite data are from three experiments, whereas protein data are from one experiment.
SEC
The SEC protocol was adjusted from Veyel et al. (2017). A total of 0.5 mL of concentrated soluble fraction corresponding to 10 mg of protein was separated by SEC. Fractionation was performed using a Superdex 200 Increase 10/300 GL prep-grade column (GE Healthcare Life Science) connected to an ÄKTA Explorer 10S (GE Healthcare Life Science) at a flow rate of 0.7 mL min−1 at 4°C. Equilibration of the column and separation were performed using 50 mm AmBIC, 150 mm NaCl, and 1.5 mm MgCl2. Twelve fractions of 1 mL were collected from an 8- to 20-mL elution volume, frozen by snap freezing in liquid nitrogen, then subsequently lyophilized and stored at −80°C for metabolite and protein extraction.
Metabolite and Protein Extraction
The protocol for the extraction of molecules was adjusted from Giavalisco et al. (2011). Small molecules and macromolecules were extracted using an MTBE/methanol/water solvent system, which separates molecules into pellet (proteins), organic (lipids), and aqueous (primary and secondary metabolites) phases. Equal volumes of particular fractions were dried using a centrifugal evaporator and stored at −80°C before metabolomics analysis.
LC-MS Metabolomics
Dried aqueous phase was measured using an ultra-performance liquid chromatograph coupled to an Exactive mass spectrometer (Thermo Fisher Scientific) in positive and negative ionization modes as described (Giavalisco et al., 2011). Processing of data was performed using REFINER MS 10.5 (GeneData) and included peak detection, chemical noise subtraction, retention time (RT) alignment, and integration of isotopic peaks into peak clusters.
Metabolite Filtering and Annotation Using an In-House Reference Compound Library
2′,3′-cAMP was annotated using an in-house reference compound library allowing 0.005-D m/z and 0.1-min RT deviations. The following criteria were applied to filter for endogenous plant mass features: present in at least two replicates with intensity threshold set to 5,000 counts and having maximum intensity in protein-containing fractions 10-fold higher than average intensity of mass features discovered as ingredients of the buffer (mobile phase of SEC, blank).
LC-MS/MS for Proteins
Protein pellets formed in the MTBE-based extraction method were solubilized in urea buffer (6 m urea and 2 m thiourea in 40 mm ammonium bicarbonate). Protein content was determined using the Bradford assay (Carl Roth). Reduction of Cys residues, alkylation, and enzymatic digestion of proteins using LysC/Trypsin Mix (Promega) were performed according to the latter’s technical manual. Peptides were desalted using C18 Empore extraction discs (3M) STAGE tips (Rappsilber et al., 2003) and dried to approximately 4 µL using a centrifugal evaporator. Samples were stored at −80°C until measurement. Dried peptides were solubilized in loading buffer (2% acetonitrile and 0.2% trifluoroacetic acid), and an equivalent of 0.8 to 1 µg of peptides was separated using a reverse-phase column and analyzed on a Q-Exactive Plus or Q-Exactive HF spectrometer (Thermo Fisher Scientific). Detailed information regarding peptide separation and identification is given in Supplemental Tables S7 to S11.
LC-MS/MS Protein Data Analysis
MaxQuant version 1.6.0.16 (Cox and Mann, 2008) and its build-in search engine Andromeda (Cox et al., 2011) were used to analyze raw proteomics data. For protein annotation, the Arabidopsis protein database, from 2014 and last updated in December 2017, was used. The search also included a contaminant database. For detailed MaxQuant settings and parameters, see Supplemental Tables S7 to S11. Contaminants and decoy hits were removed from each data set. Furthermore, at least two unique peptides were required per protein group. Label-free quantification intensities were used in all analyses performed in this study.
Data Analysis of the Time-Course Experiment
Heat and darkness Arabidopsis samples were taken from a time-course experiment performed using three biological replicates (Caldana et al., 2011). 2′,3′-cAMP and 2′,3′-cGMP were detected using an ultra-performance liquid chromatograph coupled to an Exactive mass spectrometer (Thermo Fisher Scientific) in positive and negative ionization modes as described previously (Giavalisco et al., 2011). Annotation was made allowing 0.1-min RT and 5-ppm deviations from the reference compound, supported by fragmentation data.
2′,3′-cAMP Absolute Quantification
2′,3′-cAMP reference compound (Sigma-Aldrich) was spiked into cellular extract (soluble fraction; see above) in concentrations ranging from 100 pm to 1 mm. To be able to distinguish between endogenous 2′,3′-cAMP and the reference compound, lysate was obtained from cells labeled with 15N (Kierszniowska et al., 2009). Endogenous 2′,3′-cAMP was detected at m/z = 333.03 (M-H) and the reference compound at m/z = 328.05 (M-H), the difference corresponding to five nitrogen atoms. All samples were extracted by an MTBE/methanol/water solvent system to separate proteins, lipids, and polar compounds into pellet, organic, and aqueous phases, respectively (Giavalisco et al., 2011). Samples were measured using an ultra-performance liquid chromatograph coupled to an Exactive mass spectrometer (Thermo Fisher Scientific) in positive and negative ionization modes as described previously (Giavalisco et al., 2011). Relative intensity measured for increasing concentrations of 2′,3′-cAMP standard was used to plot a calibration curve, linear in the 100 nm to 100 µm range. All measurements were done in triplicate.
Source of Recombinant Proteins
Rbp47b was cloned as a C-terminal GFP fusion (to increase protein solubility) into the Escherichia coli expression vector pDEST17 containing a His6 tag at the N terminus of the Gateway (Thermo Fisher Scientific) cassette (for vectors and oligonucleotides, see Supplemental Tables S12 and S13, respectively). Rosetta cells expressing His6-Rbp47b-GFP were grown at 28°C overnight and the next day were moved to Terrific Broth medium supplied with 1% Suc and relevant antibiotics. Cultures at OD = 0.4 were induced by the addition of 0.1 mm IPTG and transferred to 16°C for overnight incubation. The next day, cells were harvested by centrifugation, resuspended in lysis buffer (1× phosphate buffer [PBS] containing 1 mm PMSF and 1 mm imidazole), and disrupted with an Avestin EmulsiFlex C3 homogenizer. Expressed protein was purified using imidazole-gradient purification in Ni-NTA agarose (Qiagen; 34080). Next, we performed SEC (Supplemental Fig. S5) and collected 12 protein fractions. Rbp47b eluted in two fractions, first as a His6-Rbp47b-GFP fusion, second as a His6-Rbp47b fusion: spontaneous cleavage of the GFP tag is a documented phenomenon (Bird et al., 2015). His6-Rbp47b was used for MST measurements. The domains of Rbp47b, (RRM1, RRM2, and RRM3) were cloned similarly into the E. coli expression vector pDEST17. Induction and purification were performed using the protocol mentioned above, with the exception of RRM2, which was purified using Tris buffer (25 mm Tris and 0.5 m NaCl) instead. Proteins were further purified using SEC (Supplemental Fig. S11).
MST
MST measurements were performed using a Monolith NT.115 instrument (NanoTemper). Capillaries were loaded into the instrument as sets of 16 point ligand titrations. Rb47b was labeled in PBS using the Monolith protein-labeling kit RED-NHS (amine reactive; MO-L001) according to the manufacturer’s instructions. RRM1 to RRM3 were labeled using a His kit. Note that RRM2 was in Tris (25 mm Tris and 0.5 m NaCl) buffer. Excitation was optimized by varying the LED power to yield emission intensities above 200 AU, corresponding to 10 to 50 nm labeled protein. Monolith power was set to 60% to 90%. Ligands 2′,3′-cAMP (Sigma-Aldrich; A9376), 2′,3′-cGMP (Sigma-Aldrich; G8129), 3′,5′-cGMP (Sigma-Aldrich; G7504), and 3′,5′-cAMP (Sigma-Aldrich; A6885), with the exception of RRM2, were dissolved in 1× PBS. Tween (0.5%) and premium-coated capillaries were used to diminish the sticking of protein to capillaries. For the measurement of 2′,3′-cAMP-RRM domain interactions, standard capillaries were used. For the self-assembly experiment, nonlabeled Rbp47b was used as a ligand. MO Affinity Analysis software was used to analyze (Kd calculation) and visualize the data. The data presented are from three replicates (independent titrations).
nanoDSF: Protein Thermal Stability Measurements
Rbp47b and the domains were obtained as described above. 2′,3′-cAMP (2 mm) and 2′,3′-cGMP (2 mm) were prepared in 1× PBS buffer. For Rbp47b measurements, 1.5 µm protein with the addition of ligand up to 2 mm was loaded into high-sensitivity capillaries. Capillaries were loaded into Prometheus NT.48 (Nanotemper). Unfolding was detected during heating in a linear thermal ramp (2°C min−1, 20°C‒90°C) with an excitation power of 60% to 100%. Temperature-dependent protein unfolding was determined from changes in Trp and Tyr fluorescence at emission wavelengths of 350 and 330 nm. Tm values were determined by detecting the maximum of the first derivative of the fluorescence ratios (F350 nm/F330 nm) as described previously (Martin et al., 2014).
Br-2′,3′-cAMP Treatment and Confocal Microscopy
35S:GFP-Rbp47b transgenic lines used as SG markers were published by Gutierrez-Beltran et al. (2015). Five-day-old seedlings growing on vertical plates (one-half-strength Murashige and Skoog medium [Murashige and Skoog, 1962] supplemented with 1% Suc and 0.8% agar) in continuous light conditions were transferred to control medium or to medium supplemented with either 100 μm 8Br-2′,3′-cAMP (Biolog Life Science Institute) or 100 μm 8Br-3′,5′-cAMP (Sigma-Aldrich; B5386). All seedlings were first vacuum infiltrated for 5 min and then further incubated up to 1 h. As a control for SG formation, seedlings were subjected to 42°C for 30 min to 1 h. To follow the localization of the Rbp47b-GFP protein, roots were analyzed with a DM6000B/SP5 confocal laser scanning microscope (Leica Microsystems). GFP was excited with a 488-nm laser, and emission of GFP was detected between 500 and 520 nm.
Accession Number
The accession number for Rbp47b is At3g19130.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Scheme of the AP experiment using agarose beads coupled with 2′,3′-cAMP.
Supplemental Figure S2. Br-2′,3′-cAMP treatment promoted SG formation in 25% of examined seedlings.
Supplemental Figure S3. Coelution profile of the proteasome subunits in the SEC analysis presented as a heat map.
Supplemental Figure S4. Abundance of Rbp47b protein in different elutions of affinity purification experiments.
Supplemental Figure S5. Expression and purification of Rbp47b from E. coli.
Supplemental Figure S6. Rbp47b protein is stabilized in the presence of 2′,3′-cAMP.
Supplemental Figure S7. MST measurement of the interactions between Rbp47b and 3′,5′-cAMP and between Rbp47b and 3′,5′-cGMP.
Supplemental Figure S8. MST traces corresponding to Figure 4 and Supplemental Figure S7.
Supplemental Figure S9. Estimation of 2′,3′-cAMP cellular concentration in Arabidopsis native lysate (from Arabidopsis cell cultures).
Supplemental Figure S10. Amino acid sequence of the Rbp47b protein.
Supplemental Figure S11. Purification of Rbp47b domains.
Supplemental Table S1. Complete list of the proteins identified in the 2′,3′-cAMP affinity purification analysis using cell culture samples.
Supplemental Table S2. Complete list of the proteins identified in the 2′,3′-cAMP affinity purification analysis using rosette samples.
Supplemental Table S3. Complete list of the putative 2′,3′-cAMP interactors identified in the cell culture and rosette samples.
Supplemental Table S4. Complete list of the proteins more abundant (FC > 1.9) in the eluate from the heat-stressed versus control rosette samples.
Supplemental Table S5. Percentage of the 35S:GFP-Rbp47b seedlings that formed SGs in different treatments.
Supplemental Table S6. Complete list of the proteins identified in the SEC analysis and elution profiles of 2′,3′-cAMP and 2′,3′-cGMP.
Supplemental Table S7. LC-MS/MS proteomics measurements: affinity purification cell cultures.
Supplemental Table S8. LC-MS/MS proteomics measurements: size exclusion chromatography.
Supplemental Table S9. LC-MS/MS proteomics measurements: affinity purification rosettes.
Supplemental Table S10. MaxQuant output table parameters.txt: affinity purification.
Supplemental Table S11. MaxQuant output table parameters.txt: size exclusion chromatography.
Supplemental Table S12. Vectors used in this study.
Supplemental Table S13. Oligonucleotides used in this study.
Acknowledgments
We thank Lothar Willmitzer for guidance and support, Elisa Izaurralde, Dr. Lara Wohlbold, and Dr. Ruslan Nedielkov for discussing our work, and Dr. Si Wu for sharing time-course LC-MS data, used to extract 2′,3′-cAMP information. Metabolomics samples from the time-course experiment were extracted and run by Änne Michaelis. Language and scientific editing was done by Dr. Hezi Tenenboim.
Footnotes
We acknowledge the Knut and Alice Wallenberg Foundation for supporting the work of E.G.-B.
Articles can be viewed without a subscription.
References
- Anderson P, Kedersha N (2008) Stress granules: the Tao of RNA triage. Trends Biochem Sci 33: 141–150 [DOI] [PubMed] [Google Scholar]
- Arimoto-Matsuzaki K, Saito H, Takekawa M (2016) TIA1 oxidation inhibits stress granule assembly and sensitizes cells to stress-induced apoptosis. Nat Commun 7: 10252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryal UK, Xiong Y, McBride Z, Kihara D, Xie J, Hall MC, Szymanski DB (2014) A proteomic strategy for global analysis of plant protein complexes. Plant Cell 26: 3867–3882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aye TT, Mohammed S, van den Toorn HW, van Veen TA, van der Heyden MA, Scholten A, Heck AJ (2009) Selectivity in enrichment of cAMP-dependent protein kinase regulatory subunits type I and type II and their interactors using modified cAMP affinity resins. Mol Cell Proteomics 8: 1016–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azarashvili T, Krestinina O, Galvita A, Grachev D, Baburina Y, Stricker R, Evtodienko Y, Reiser G (2009) Ca2+-dependent permeability transition regulation in rat brain mitochondria by 2′,3′-cyclic nucleotides and 2′,3′-cyclic nucleotide 3′-phosphodiesterase. Am J Physiol Cell Physiol 296: C1428–C1439 [DOI] [PubMed] [Google Scholar]
- Baginsky S, Gruissem W (2002) Endonucleolytic activation directs dark-induced chloroplast mRNA degradation. Nucleic Acids Res 30: 4527–4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard C, Traub M, Kunz HH, Hach S, Trentmann O, Möhlmann T (2011) Equilibrative nucleoside transporter 1 (ENT1) is critical for pollen germination and vegetative growth in Arabidopsis. J Exp Bot 62: 4627–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird LE, Rada H, Verma A, Gasper R, Birch J, Jennions M, Lӧwe J, Moraes I, Owens RJ (2015) Green fluorescent protein-based expression screening of membrane proteins in Escherichia coli. J Vis Exp e52357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Parker R (2009) Eukaryotic stress granules: the ins and outs of translation. Mol Cell 36: 932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldana C, Degenkolbe T, Cuadros-Inostroza A, Klie S, Sulpice R, Leisse A, Steinhauser D, Fernie AR, Willmitzer L, Hannah MA (2011) High-density kinetic analysis of the metabolomic and transcriptomic response of Arabidopsis to eight environmental conditions. Plant J 67: 869–884 [DOI] [PubMed] [Google Scholar]
- Chantarachot T, Bailey-Serres J (2018) Polysomes, stress granules, and processing bodies: a dynamic triumvirate controlling cytoplasmic mRNA fate and function. Plant Physiol 176: 254–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10: 1794–1805 [DOI] [PubMed] [Google Scholar]
- Culver GM, Consaul SA, Tycowski KT, Filipowicz W, Phizicky EM (1994) tRNA splicing in yeast and wheat germ: a cyclic phosphodiesterase implicated in the metabolism of ADP-ribose 1′′,2′′-cyclic phosphate. J Biol Chem 269: 24928–24934 [PubMed] [Google Scholar]
- Donaldson L, Meier S, Gehring C (2016) The Arabidopsis cyclic nucleotide interactome. Cell Commun Signal 14: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupeux F, Santiago J, Betz K, Twycross J, Park SY, Rodriguez L, Gonzalez-Guzman M, Jensen MR, Krasnogor N, Blackledge M, et al. (2011) A thermodynamic switch modulates abscisic acid receptor sensitivity. EMBO J 30: 4171–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik P, Hall J, Filipowicz W (1997) Cloning and characterization of the Arabidopsis cyclic phosphodiesterase which hydrolyzes ADP-ribose 1′′,2′′-cyclic phosphate and nucleoside 2′,3′-cyclic phosphates. J Biol Chem 272: 13211–13219 [DOI] [PubMed] [Google Scholar]
- Giavalisco P, Li Y, Matthes A, Eckhardt A, Hubberten HM, Hesse H, Segu S, Hummel J, Köhl K, Willmitzer L (2011) Elemental formula annotation of polar and lipophilic metabolites using 13C, 15N and 34S isotope labelling, in combination with high-resolution mass spectrometry. Plant J 68: 364–376 [DOI] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P (2004) Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell 15: 5383–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Beltran E, Moschou PN, Smertenko AP, Bozhkov PV (2015) Tudor staphylococcal nuclease links formation of stress granules and processing bodies with mRNA catabolism in Arabidopsis. Plant Cell 27: 926–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann A, Zdanov A, Genschik P, Ruvinov S, Filipowicz W, Wlodawer A (2000) Structure and mechanism of activity of the cyclic phosphodiesterase of Appr>p, a product of the tRNA splicing reaction. EMBO J 19: 6207–6217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EK. (2017) Discovery and roles of 2′,3′-cAMP in biological systems. Handb Exp Pharmacol 238: 229–252 [DOI] [PubMed] [Google Scholar]
- Jackson EK, Ren J, Mi Z (2009) Extracellular 2′,3′-cAMP is a source of adenosine. J Biol Chem 284: 33097–33106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R (2016) ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164: 487–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerabek-Willemsen M, Wienken CJ, Braun D, Baaske P, Duhr S (2011) Molecular interaction studies using microscale thermophoresis. Assay Drug Dev Technol 9: 342–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P (1999) RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol 147: 1431–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A, Matheny T, Jain S, Mitchell SF, Wheeler JR, Parker R (2017) The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol Cell 68: 808–820.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierszniowska S, Walther D, Schulze WX (2009) Ratio-dependent significance thresholds in reciprocal 15N-labeling experiments as a robust tool in detection of candidate proteins responding to biological treatment. Proteomics 9: 1916–1924 [DOI] [PubMed] [Google Scholar]
- Lappe-Siefke C, Goebbels S, Gravel M, Nicksch E, Lee J, Braun PE, Griffiths IR, Nave KA (2003) Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat Genet 33: 366–374 [DOI] [PubMed] [Google Scholar]
- Lechler MC, David DC (2017) More stressed out with age? Check your RNA granule aggregation. Prion 11: 313–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Wei HM, Chen LY, Li C (2014) Localization of SERBP1 in stress granules and nucleoli. FEBS J 281: 352–364 [DOI] [PubMed] [Google Scholar]
- Li CH, Ohn T, Ivanov P, Tisdale S, Anderson P (2010) eIF5A promotes translation elongation, polysome disassembly and stress granule assembly. PLoS ONE 5: e9942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorković ZJ, Wieczorek Kirk DA, Klahre U, Hemmings-Mieszczak M, Filipowicz W (2000) RBP45 and RBP47, two oligouridylate-specific hnRNP-like proteins interacting with poly(A)+ RNA in nuclei of plant cells. RNA 6: 1610–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin L, Schwarz S, Breitsprecher D (2014) Analyzing Thermal Unfolding of Proteins: The Prometheus NT.48. Application Note NT-PR-001 [Google Scholar]
- Menges M, Murray JA (2002) Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J 30: 203–212 [DOI] [PubMed] [Google Scholar]
- Merret R, Descombin J, Juan YT, Favory JJ, Carpentier MC, Chaparro C, Charng YY, Deragon JM, Bousquet-Antonelli C (2013) XRN4 and LARP1 are required for a heat-triggered mRNA decay pathway involved in plant acclimation and survival during thermal stress. Cell Rep 5: 1279–1293 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nover L, Scharf KD, Neumann D (1989) Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol Cell Biol 9: 1298–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Ishihama Y, Mann M (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 75: 663–670 [DOI] [PubMed] [Google Scholar]
- Ren J, Mi Z, Stewart NA, Jackson EK (2009) Identification and quantification of 2′,3′-cAMP release by the kidney. J Pharmacol Exp Ther 328: 855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorenson R, Bailey-Serres J (2014) Selective mRNA sequestration by OLIGOURIDYLATE-BINDING PROTEIN 1 contributes to translational control during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 111: 2373–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Santos A, von Mering C, Jensen LJ, Bork P, Kuhn M (2016) STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res 44: D380–D384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JE, Venegas FD, Raines RT (1994) Energetics of catalysis by ribonucleases: fate of the 2′,3′-cyclic phosphodiester intermediate. Biochemistry 33: 7408–7414 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, et al. (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Van Damme T, Blancquaert D, Couturon P, Van Der Straeten D, Sandra P, Lynen F (2014) Wounding stress causes rapid increase in concentration of the naturally occurring 2′,3′-isomers of cyclic guanosine- and cyclic adenosine monophosphate (cGMP and cAMP) in plant tissues. Phytochemistry 103: 59–66 [DOI] [PubMed] [Google Scholar]
- Verrier JD, Jackson TC, Bansal R, Kochanek PM, Puccio AM, Okonkwo DO, Jackson EK (2012) The brain in vivo expresses the 2′,3′-cAMP-adenosine pathway. J Neurochem 122: 115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyel D, Kierszniowska S, Kosmacz M, Sokolowska EM, Michaelis A, Luzarowski M, Szlachetko J, Willmitzer L, Skirycz A (2017) System-wide detection of protein-small molecule complexes suggests extensive metabolite regulation in plants. Sci Rep 7: 42387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel US, Thompson RJ (1987) Molecular cloning of the myelin specific enzyme 2′,3′-cyclic-nucleotide 3′-phosphohydrolase. FEBS Lett 218: 261–265 [DOI] [PubMed] [Google Scholar]
- Weber C, Nover L, Fauth M (2008) Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J 56: 517–530 [DOI] [PubMed] [Google Scholar]
- Wheeler JR, Matheny T, Jain S, Abrisch R, Parker R (2016) Distinct stages in stress granule assembly and disassembly. eLife 5: e18413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Yan Z, Wang Y, Yan X, Han Y (2014) Tudor-SN, a component of stress granules, regulates growth under salt stress by modulating GA20ox3 mRNA levels in Arabidopsis. J Exp Bot 65: 5933–5944 [DOI] [PMC free article] [PubMed] [Google Scholar]