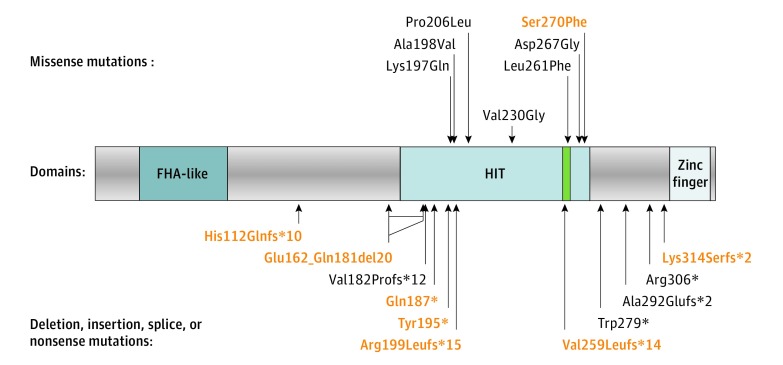
Figure 1. Position of the Mutations With Respect to the Aprataxin (APTX) Protein Domains.
The histidine triad (HIT) motif (HxHxH) is highlighted in green. The deletion, insertion, and splice site mutations are translated into estimated protein consequences. Correspondence for novel mutations (red) is indicated in Table 3; c.544-2A>G (acceptor) and c.770 + 1G>A (donor) exon 5 splice site mutations are estimated to result in frameshift exon 5 skipping p.Val182Profs*12; c.875-1G>A (last) exon 7 acceptor splice site mutation is estimated to result in use of a minor alternative exon 7’ (isoform coded by transcript NM_175069) Ala292Glufs*2. The position of the in-frame exon 4 deletion (Glu162_Gln181del20) is indicated by a double arrow that represents the extent of internal peptide deletion. The large deletion of exons 1 to 4 is not represented because it is estimated to result in complete absence of APTX transcription.