Figure 6.
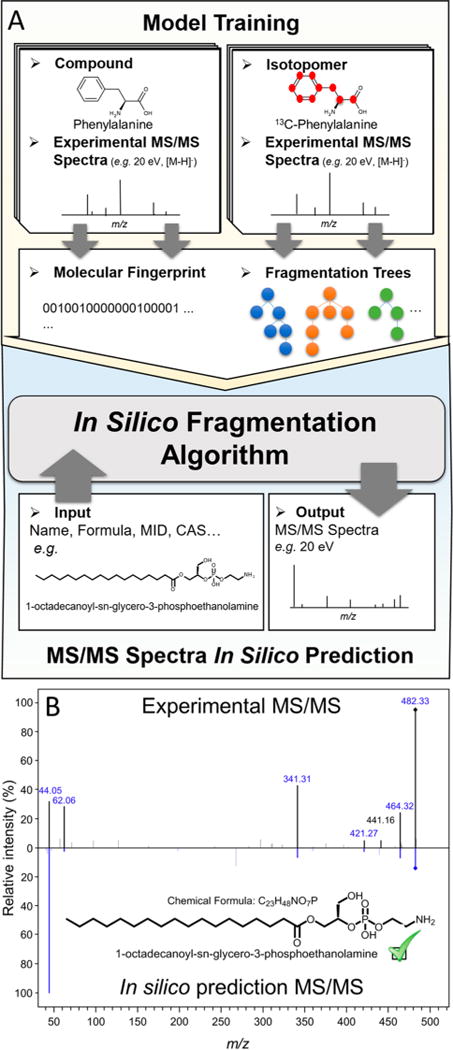
In silico data generation. (A) Workflow for in silico data simulation. A generalization of the input–output kernel regression model, especially designed to predict fragments of known molecules, is used to generate in silico data. Both unlabeled and isotope-labeled compounds are used for model training, providing additional information through the number of isotope-labeled atoms of each fragment. (B) Comparison between experimental MS/MS spectrum generated by lysoPE(18:0) with its in silico prediction in METLIN, at a collision energy of 10 eV. It is worth noting that 6 out of 7 main fragments of the experimental spectrum match with the in silico simulated data (highlighted in blue).
