Figure 3. U2AF1 and SRSF2 Mutations in the Pancan Cohort and Differential Splicing Associated with Hotspot Mutations.
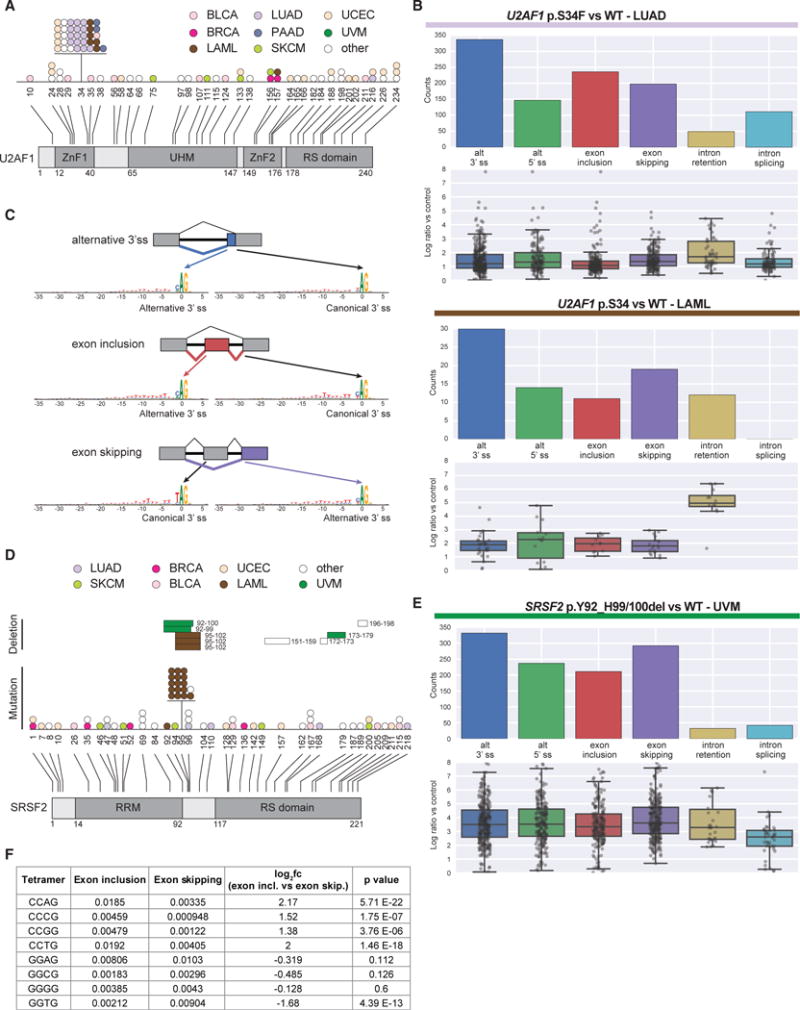
(A) U2AF1 somatic mutations mapped to amino acid positions and annotated domains. Each dot represents a single mutant sample colored by tumor cohort.
(B) Differential splicing events associated with U2AF1 p.S34F/Y hotspot mutations in LUAD and LAML (corrected q-value < 0.05). Below each splicing event count, the PSI log2 fold change of each individual event is detailed in a boxplot.
(C) Consensus sequence motifs for exons preferentially used by mutant U2AF1 versus WT U2AF1 across various alternative splicing events.
(D) SRSF2 somatic mutations mapped to amino acid positions and annotated domains. Each bar (in-frame deletion) and dot (other mutations) represents a singlemutant sample colored by tumor cohort.
(E) Differential splicing events associated with SRSF2 in-frame deletions in UVM.
(F) Tetramer (CCNG and GGNG) enrichment analysis comparing cassette exons preferentially included or excluded by SRSF2 mutant samples. Each value is the average tetramer occurrence frequency for all exons in that class. Fold change significance was assessed using Student’s t test.
See also Tables S2 and S3.
