Figure 6. Pathway Enrichment Analysis Using Curated Gene Sets Indicates that Cancer Pathways Altered by Splicing Factor Mutations Are Lineage Specific.
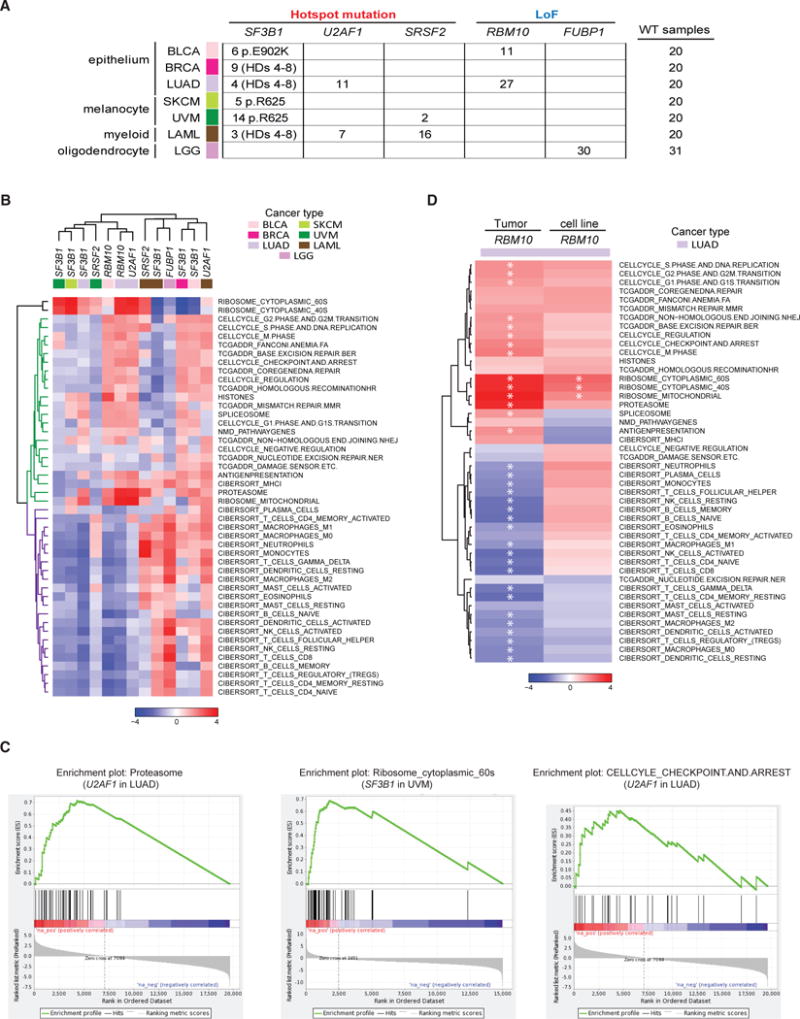
(A) Splicing factor gene mutations and their associated tumor cohorts used in pathway analyses.
(B) Heatmap of gene set enrichment analyses for all comparison groups generated using normalized enrichment scores (NESs) of 46 curated gene sets. Two distinct subclasses of gene sets are cell-autonomous pathways (green) and immune-related signatures (purple).
(C) Representative cancer hallmark gene sets upregulated in splicing factor mutant samples.
(D) Heatmap of NESs comparing patient tumor samples and cell lines, where each column represents the differential pathway modulation of RBM10 LoF mutants (n = 27 TCGA, n = 3 cell lines) versus RBM10 WT (n = 20 TCGA, n = 30 cell lines) of 46 curated gene sets. Significantly modulated gene sets (q value ≤ 0.05) are highlighted with an asterisk.
See also Figure S5 and Tables S4, S5, S6, and S7.
