Abstract
Here, the term “module” is redefined for trans-acyltransferase (trans-AT) assembly lines to agree with how its domains cooperate and evolutionarily co-migrate. The key domain in both the polyketide synthase (PKS) and nonribosomal peptide synthetase (NRPS) modules of assembly lines is the acyl carrier protein (ACP). ACPs not only relay growing acyl chains through the assembly line but also collaborate with enzymes in modules, both in cis and in trans, to add a specific chemical moiety. A ketosynthase (KS) downstream of ACP often plays the role of gatekeeper, ensuring that only a single intermediate generated by the enzymes of a module is passed downstream. Bioinformatic analysis of 526 ACPs from 33 characterized trans-AT assembly lines reveals ACPs from the same module type generally clade together, reflective of the co-evolution of these domains with their cognate enzymes. While KSs downstream of ACPs from the same module type generally also clade together, KSs upstream of ACPs do not – in disagreement with the traditional definition of a module. Beyond nomenclature, the presented analysis impacts our understanding of module function, the evolution of assembly lines, pathway prediction, and assembly line engineering.
Keywords: Polyketide, polyketide synthase, trans-AT assembly line, bioinformatics, acyl carrier protein, module redefinition
Introduction
Acyl carrier proteins (ACPs) form transient domain-domain interactions with cognate enzymes in their biosynthetic pathways1–2. These interactions enable specific transformations to occur on the acyl chain covalently shuttled by that ACP. This holds for both Type II systems in which each domain is encoded on a separate polypeptide, as in most bacterial fatty acid synthases (FASs)3–4, as well as Type I systems in which several domains are encoded on the same polypeptide, as in biosynthetic assembly lines formed by polyketide synthase (PKS)5–6 and nonribosomal peptide synthetase (NRPS) machinery7.
The repeating unit of enzymatic assembly lines is known as the “module”. The domains within a module typically fall into one of four categories: 1) an enzyme that selects and transfers a building block to an ACP, 2) an ACP that covalently shuttles a building block and/or an acyl chain, 3) a processing enzyme that performs chemistry on the acyl chain, or 4) a chain extending/transferring/releasing (ETR) enzyme. Generally, modules extend an acyl chain with a carboxylic acid building block selected by an acyltransferase (AT) or an amino acid building block selected by an adenylation (A) domain. An ACP transporting an α-carboxyacyl group usually collaborates with an upstream ketosynthase (KS) domain to generate a carbon-carbon bond and extend the acyl chain, while an ACP transporting an α-aminoacyl group [a.k.a. peptidyl carrier protein (PCP) or thiolation (T) domain] usually collaborates with an upstream condensation (C) domain to generate a carbon-nitrogen bond and extend the acyl chain.
The biosynthetic community is working to decipher the logic of two large classes of assembly lines that contain PKS machinery - cis-AT assembly lines that primarily rely on embedded ATs5, 8 and trans-AT assembly lines that primarily rely on separately-encoded ATs6, 9. From the sequencing of the erythromycin synthase in 1990 until last year, the boundaries of the modules of cis-AT assembly lines were incorrectly defined - comparisons with the domain organization of the mammalian FAS had led to the module being defined with KS at its upstream boundary and ACP at its downstream boundary. The processing enzymes in several cis-AT assembly lines have been shown to evolutionarily co-migrate with the KS downstream of them, leading to the redefinition of cis-AT modules as possessing a downstream KS5, 10. When trans-AT assembly lines started to be discovered fifteen years ago, often from difficult-to-culture, symbiotic bacteria, the use of cis-AT nomenclature resulted in their modules being incorrectly defined as well11–12. Only a few years later KSs that accept similar substrates in trans-AT assembly lines were oberved to clade together, suggesting that they collaborate most closely with the domains preceding them, not following them13.
Bioinformatic analysis of KS domains has helped predict the intermediates and products of trans-AT assembly lines13 (analyzing which activities are embedded in the assembly line is insufficient due to the common participation of trans-enzymes). Because the KSs of trans-AT assembly lines often serve as gatekeepers, the clade to which a KS belongs can suggest the chemical moiety most recently added to the acyl chain. However, relying on KS signatures alone has limitations since KS clades do not by themselves always reveal the chemistry of the acyl chain and KSs are often replaced by other ETR enzymes, such as a TE or C domain. To better predict products and decipher how the many module types of trans-AT assembly lines operate, all the domains of the module should be considered. An examination of trans-AT assembly lines in light of the updated module definition, reveals that ACP sequences can help determine the module types present in an assembly line (Figure 1).
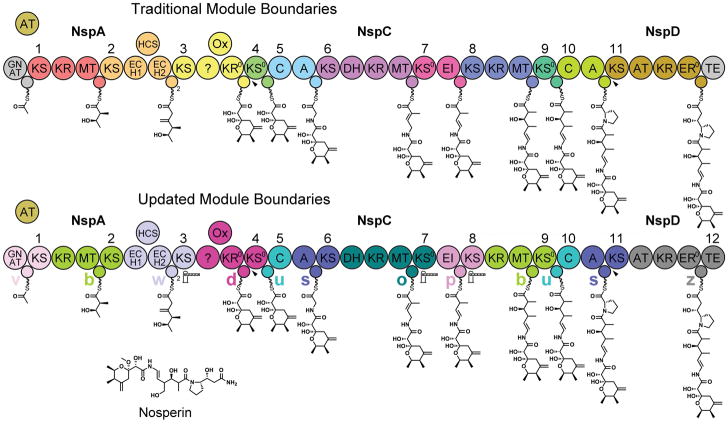
Figure 1. Traditional and Updated Module Boundaries for a trans-AT Assembly Line.
The nosperin trans-AT assembly line can be depicted either with traditional modules that start with a KS or C domain and end with an ACP domain or with updated modules that contain a central ACP domain and a downstream extending/transferring/releasing (ETR) enzyme (KS, KS0, C, or TE). All but the twelfth module of the nosperin assembly line belong to the module types defined here (a–y). Gatekeeping ETR enzymes that select one intermediate over another and disconnections between assembly line polypeptides are indicated with gate icons and arrowheads, respectively.
That little research on the bioinformatics of the ACP domains of assembly lines has been reported is understandable at least in cis-AT assembly lines where ACPs appear quite homogenous. One study did uncover a signature in the ACPs of both cis-AT assembly lines (curacin and jamaicamide) and trans-AT assembly lines (bacillaene, batumin, mupirocin, pederin, and thiomarinol) that flags their β-ketoacyl substrates for β-branching reactions14. In each case, the acyl-ACP must associate with a hydroxymethyl-CoA synthase like enzyme (HCS) and two enoyl-CoA hydrolase like enyzmes (ECH1 and ECH2). Presumably, the signature is representative of unique features on the ACP surface that enable it to selectively associate with these enzymes.
Here, bioinformatic analysis of 526 ACPs from 33 well-characterized trans-AT assembly lines reveals that ACPs from β-branching modules are only one of several groups of ACPs from trans-AT assembly lines that clade together. Indeed, most ACPs from the same module type were found to be related to one another, reflective of their interactions with cognate processing enzymes that have been relatively conserved through evolution. KSs downstream of ACPs from the same module type also clade together, whereas KSs upstream do not. Beyond nomenclature, how the presented analysis impacts our understanding of modules, the evolution of assembly lines, pathway prediction, and assembly line engineering is discussed.
Materials and Methods
Well-characterized, nonredundant trans-AT assembly lines with reasonable biosynthetic models were chosen for analysis (albicidin, Alb, BGC0001088; bacillaene, Bae, BGC0001089; basiliskamides, Bas, BGC0000172; bongkrekic acid, Bon, BGC0000173; bryostatin, Bry, BGC0000174; calyculin, Cal, BGC0000967; chivosazole, Chi, BGC0001069; corallopyronin, Cor, BGC0001091; difficidin, Dif, BGC0000176; disorazole, Dsz, BGC0001093; migrastatin, Mgs, BGC0000177; elansolid, Ela, BGC0000178; enacyloxin, Ena, BGC0001094; etnangien, Etn, BGC0000179; griseoviridin, Sgv, BGC0000459; batumin, Bat, BGC0001099; kirromycin, Kir, BGC0001070; legioliulin, Lgl, BGC0000180; leinamycin, Lnm, BGC0001101; macrolactin, Mln, BGC0000181; mupirocin, Mmp, BGC0000182; myxovirescin, Myx, BGC0001025; nosperin, Nsp, BGC0001071; oocydin, Ooc, BGC0001031; onnamide, Onn, BGC0001105; oxazolomycin, Ozm, BGC0001106; patellazole, Ptz, BGC0001107; psymberin, Psy, BGC0001110; rhizopodins, Riz, BGC0001111; rhizoxins, Rhi, BGC0001112; sorangicin, Sor, BGC0000184; thailandamide, Tai, BGC0000186, and thailanstatin, Tst, BGC0001114). Modules from cis-AT assembly lines were also selected for comparison (amphotericin module 10, Amp10, BGC0000015; concanamycin module 6, Con6, BGC0000040; erythromycin module 4, Ery4, BGC0000055; mycolactone module 4, Myc4, BGC0000103; nystatin module 9, Nys9, BGC0000115; pikromycin module 5, Pik5, BGC0000094; rapamycin module 6, Rap6, BGC0001040; rifamycin module 5, Rif5, BGC0000136; spinosad module 5, Spn5, BGC0000148; and tylosin module 3, Tyl3, BGC0000166)6, 9, 15.
All presented cladograms were similarly generated. First, sequences were aligned with MUSCLE16. Then RAxML performed rapid bootstrapping and a subsequent maximum-likelihood (ML) search on each alignment17. To construct trees, the LG amino acid substitution matrix as well as the GAMMA model of rate heterogeneity were utilized and the alpha-parameter was estimated by ML18. The random seed for parsimony inferences and for rapid bootstraps (500 for ACPs, 100 for KSs) was 22222. ACP and KS trees were rooted by representative domains from cis-AT assembly lines. Finally, cladograms were visualized using Dendroscope 3, with confidence intervals ≥60 displayed19.
Results and Discussion
Module Types and ACP Families Defined
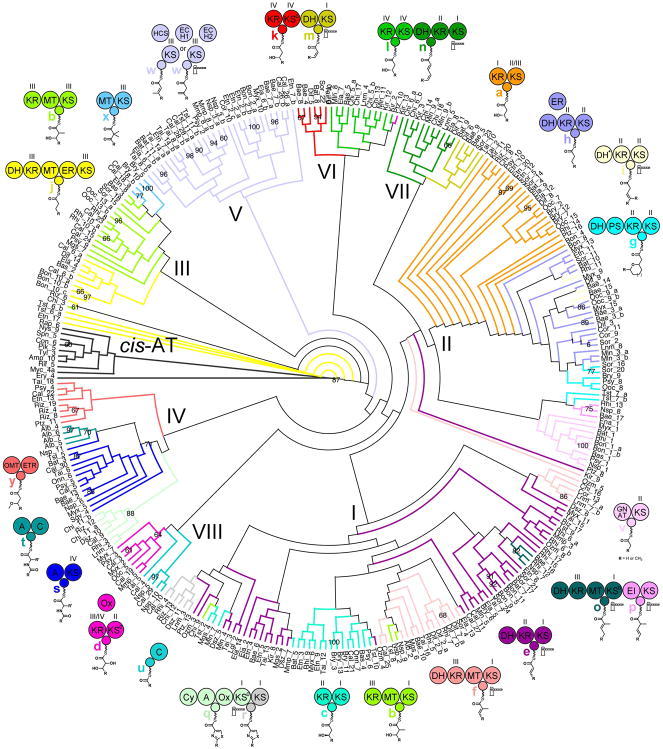
To color ACPs in the ACP cladogram by the type of module to which they belong a definition for “module type” was developed. Defining modules only by the chemistry they perform does not sufficiently explain the clading observed in the ACP cladogram (Figure S1). Including the classes of enzymes they contain in the definition significantly helps. However, to best explain ACP clading, how ACPs and their cognate enzymes have co-evolved to interact with one another also needs to be considered. Since ACPs that belong to the same major divisions of the cladogram (families) are likely to be evolutionarily related, module types were analyzed in the context of each ACP family. Thus, each of Families I-VIII contain a different set of module types (a–z, with z representing all undefined module types). A cladogram of ACPs that belong to the defined module types was generated and colored by those module types (Figure 2).
Figure 2. Cladogram of ACPs Colored by Module Type.
An analysis of the ACPs from 33 trans-AT assembly lines and 10 ACPs from cis-AT assembly lines revealed that ACPs from the same types of modules clade together and helped to define the most common module types (Figure S1). Shown here is the cladogram of the ACPs from twenty-five module types (a–y), characterized by the moiety they add as well as the classes of enzyme they contain (KSs and KRs are divided into four clades as in Figures S2 & S3). ACP families (I–VIII) that contain related module types were also defined. Suspected gatekeeping enzymes are conservatively indicated with gate icons.
Within trans-AT assembly lines, ACPs most commonly collaborate with KSs and ketoreductases (KRs) that themselves are divided into clades. From cladograms of KSs in the analyzed assembly lines, four clades were defined similar to a previous analysis (Figure S2)20. The KSI clade contains KSs that gatekeep for Cα/Cβ-double bonds. The KSII clade contains KSs that gatekeep for Cα/Cβ-single bonds. With the exception of KSs in the KSIII clade associated with β-branching, members of the KSIII and KSIV clades generally do not appear to gatekeep. The structural biology of KSs supports these clade divisions (KSI, MgsKS6, PDB 4TKT; KSII, PksKS2, PDB 4NA1; KSIV, OzmKS9, PDB 4OPF)20–21. From a cladogram of KRs from the defined module types, four main clades were observed (Figure S3). The KRI clade contains KRs that are from methyltransferase (MT)-less modules and generally A-type (A-type KRs generate L-β-hydroxy groups), the KRII contains KRs that are from MT-less modules and generally B-type (B-type KRs generate D-β-hydroxy groups), the KRIII clade contains KRs that are from MT-containing modules and can be A-type or B-type, and the KRIV clade contains KRs that are from the first module of dehydrating bimodules as well as some α-oxidizing modules and are A-type. The structural biology of KRs supports these clade divisions (KRII, MlnKR7, PDB 5D2E; KRIV, PksKR4, PDB 5KTK)22–24.
ACPs Evolutionarily Co-migrate with Downstream KSs, not Upstream KSs
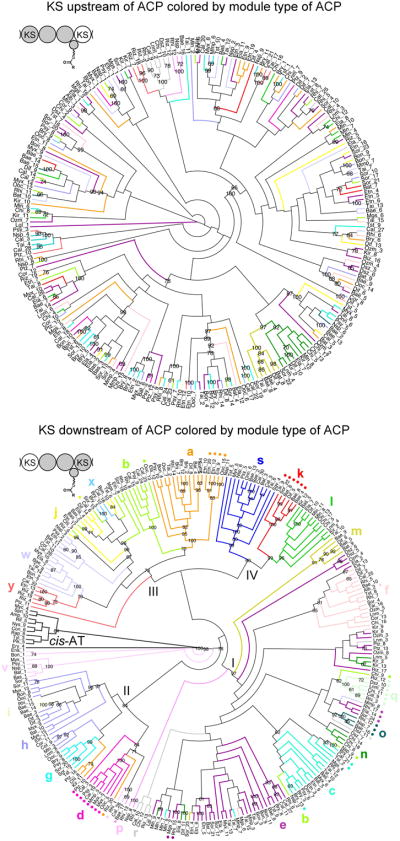
Cladograms were generated for the KSs downstream and upstream of ACPs that were assigned a module type (Figure 3). In the cladogram of downstream KSs, KSs were colored by the module type of the ACP they follow, and in the cladogram of upstream KSs, KSs were colored by the module type of the ACP following them. From this analysis it is apparent that KSs downstream of ACPs from the same module type clade with one another and that KSs upstream of ACPs from the same module type do not, with the expected exception of those in the first module of a bimodule13. This analysis supports the redefinition of trans-AT PKS modules such that ACPs are grouped with the KSs downstream of them5, 10.
Figure 3. Cladograms of KSs Upstream and Downstream of ACPs Support the Updated Module Boundaries.
A) The lack of clustering for KSs upstream of ACPs that clade together, with the anticipated exception of KSs from the first module of bimodules, indicates ACPs do not usually evolutionarily co-migrate with the KS upstream of them. B) Clustering is observed for KSs downstream of ACPs that clade together. This observation suggests that ACPs evolutionarily generally co-migrate with the KS downstream of them, reaffirming the updated module boundaries. Colored dots indicate condensation-incompetent KS0s.
The collaboration of processing enzymes with a downstream KS (or other ETR enzyme) is often required to ensure that a single acyl intermediate is passed to the next module in the assembly line. While processing reactions driven by the high NADPH/NADP+ and SAM/SAH ratios in the cell [e.g., those mediated by KR, enoylreductase (ER), and MT], essentially go to completion given a chance, others are in an equilibrium such that an ACP presents more than one acyl chain to the KS downstream of it. Thus, KSIs play a gatekeeping role in modules that generate double bonds, accepting an α/β-unsaturated intermediate but not the more abundant β-hydroxy intermediate [interconverted by a dehydratase (DH) in these modules]. KSIIs play a gatekeeping role in modules that generate β/γ-unsaturated intermediates from α/β-unsaturated intermediates [through a DH*; KSs from a separate clade similarly gatekeep in the second module of enoyl-isomerase (EI) bimodules]25. Some KSIIIs in β-branching modules apparently gatekeep to select an intermediate with a β-exomethylene group over one with an α/β-olefin. In this work, gate icons have been conservatively placed next to ETR enzymes suspected of gatekeeping.
The abundance of condensation-incompetent KSs, termed KS0s (indicated by colored dots in Figure 3; usually these possess deviations in the HGTGT motif that in condensation-competent KSs contains the first histidine of the Cys/His/His triad), in trans-AT assembly lines illustrates that KSs have several important roles besides forming carbon-carbon bonds. Viewed from the perspective of cis-AT assembly lines that are comprised of very few KS0s, these domains seemingly perform unnecessary reactions, transferring the acyl chain from one ACP to another through an acyl-KS0 intermediate. However, KS0s can play a gatekeeping role and/or transfer acyl chains between different types of ACP such that the chain can be operated on by other processing or ETR enzymes. Catalytically-competent KSs can also perform these functions but mandate a chain extension; thus, KS0s also provide chemical flexibility to trans-AT assembly lines.
ACP Sequence Motifs and Structure
To better understand how ACPs flag their cognate enzymes, sequence alignments and sequence logos were generated for ACPs of each of the common module types (Figures 4 and S4)26. As noticed with ACPs of β-branching modules (Family V)14, sequence motifs for many of the module types are evident. To obtain a better physical understanding of the motifs, the structural biology of assembly line ACPs was investigated.
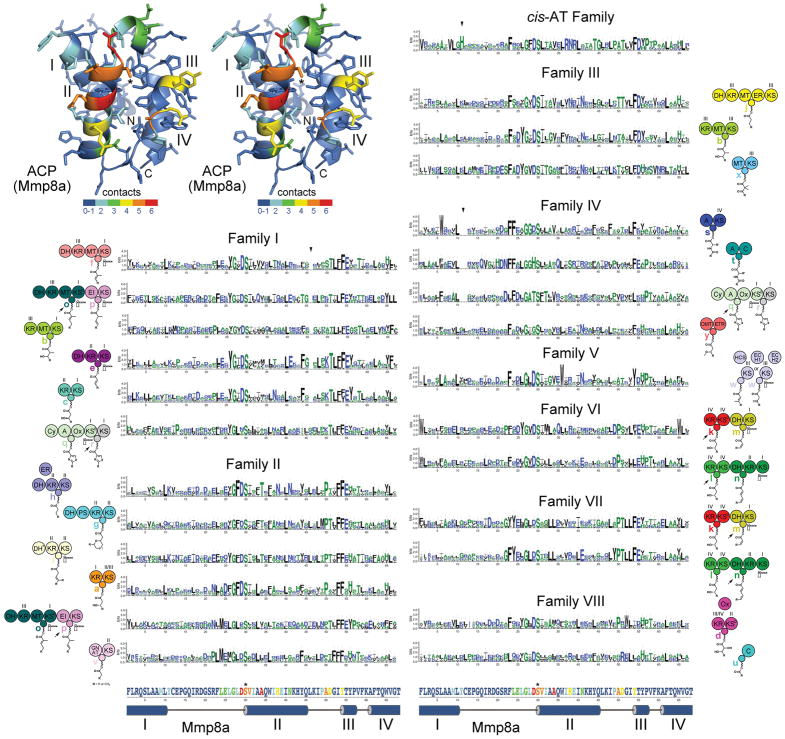
Figure 4. ACP Signatures.
A stereodiagram of an ACP (Mmp8a from the mupirocin trans-AT assembly line, PDB 2L22) shows which positions in assembly line ACPs most frequently contact cognate enzymes in the six available complex structures (red indicates that in 6/6 structures a residue in that position is within 4 Å of the cognate enzyme). The four helices (I–IV), the phosphopantetheinylated serine (*), and the boundaries of the ACP domain (N and C) are marked. Sequence logos show the ACP signatures for each of the module types, which are indicated by the neighboring cartoon. Triangles indicate relative gaps or insertions: in Family I the loop between helices II and III is generally one residue shorter, in Family IV the loop between helices I and II is generally two residues shorter, in cis-AT ACPs the loop between helices I and II is generally one residue longer and contains a conserved histidine. The primary and secondary structure of Mmp8a at the bottom helps show where residues in the ACP signatures are structurally positioned and how likely they are to make contact with cognate enzymes.
Many examples of assembly line ACPs docking with cognate enzymes are known, even though all are from NRPS modules. Representative interactions between ACPs and embedded assembly line enzymes have been observed for the C domain (donor site: PDB 5EJD27, acceptor site: PDB 4ZXH28), the A domain (PDBs 4DG929), the epimerization (E) domain (PDB 5ISX30), and the thioesterase (TE) domain (PDB 3TEJ31). Representative interactions between an ACP and a non-embedded P450 monooxygenase have also been observed (PDB 4PXH32). With these six complexes an interaction map was constructed (Figure 4). Residues on a structurally-elucidated ACP from the mupirocin trans-AT assembly line (MmpACP8a, PDB 2L22)14 were colored from blue to red (with red representing that in 6 out of 6 structures the equivalent residue is within 4 Å of the cognate enzyme). While each interaction with a cognate enzyme is unique, the majority of contacts are made with ACP helices II and III. No structures of assembly line ACPs interacting with the enzymes of PKS modules are available; however, the VinK/VinL complex (ATL and ACPL from the vicenistatin cis-AT assembly line) may be representative of how ACPs and ATs associate (PDB 5CZD)33.
While significant differences between module types are apparent, the largest distinctions occur between families (Figure 4). Compared to the average ACP, most ACPs in Family I contain one fewer residue in the loop connecting helices II and III, and most ACPs in Family IV contain two fewer residues in the loop connecting helices I and II. The loop connecting helices I and II is one residue longer in ACPs from cis-AT modules and contains a highly conserved histidine. On the surface, b modules from Family I and Family III appear similar, containing KR+MT+ACP+KS architectures and generating α-methyl, β-hydroxy moieties; however, their ACPs possess different signatures that likely reflect the distinct interfaces that each ACP forms with its cognate KR and MT.
Multiple ACPs in the same module, called tandem ACPs, are thought to increase the flux through an otherwise rate-limiting portion of the assembly line34. This is evident in β-branching modules that typically contain two to three ACPs - for each of the individual ACPs to interact with HCS, ECH1, and ECH2 as well as AT, an upstream KS, and an ETR enzyme, they possess very similar sequences. In other types of modules, tandem ACPs are also highly homologous, appearing adjacent to one another in the ACP cladogram (Figure 2). Even ACPs separated by an enzymatic domain, like the two ACPs in the seventh module of the leinamycin assembly line located before and after an MT, are highly homologous. Divergent sequences of ACPs within the same module indicate they play different roles. The two ACPs in the termination module of the leinamycin assembly line apparently help with different steps in the construction of a 1,3-dioxo-1,2-dithiolane moiety35. The two ACPs that appear to belong to the ninth module of the oxazolomycin assembly line actually interact with different downstream KSs, as discussed in the next section.
Pathway Prediction
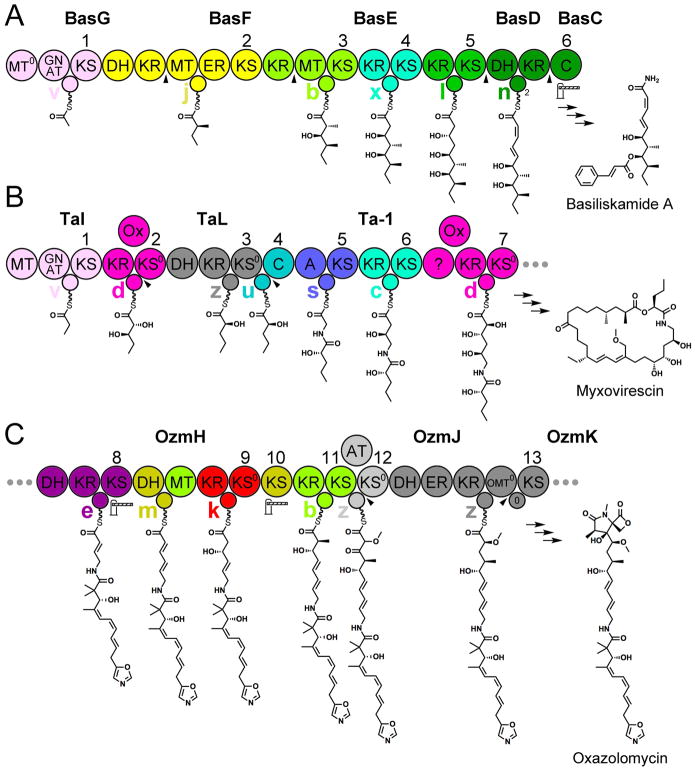
The generation of biosynthetic models for trans-AT assembly lines, such as the basiliskamide, myxovirescin, and oxazolomycin is greatly facilitated by determining the module type to which its ACPs belong (Figure 5).
Figure 5. Pathway Prediction.
Three examples show how the new understanding of trans-AT modules can be employed to generate better biosynthetic models. A) The order and function of the basiliskamide polypeptides had been mysterious. Assembling them to generate module types defined here enabled the unambiguous ordering BasG, BasF, BasE, BasD, BasC. An unusual feature of the assembly line is the final C domain that apparently gatekeeps for an α/β-cis, γ/δ-trans diene, in contrast to KSs in its position that usually gatekeep for the α/β-trans, γ/δ-cis diene, but like the similarly-positioned macrolactin TE. B) The order and function of the first polypeptides (TaI, TaL, and Ta-1) of the myxovirescin pathway had also been cryptic. Assembling the polypeptides to form module types defined here, the order could be confidently determined. The first v module generates a propionyl group, possibly through the methylation and decarboxylation of a malonyl group. The second d module reduces the β-carbon and oxidizes the α-carbon. The third module is an uncategorized module type (z) that likely generates an α-hydroxy group through a DH-mediated dehydration to an α-keto group followed by KR-mediated α-ketoreduction similar to PksKR3 (Ref. 22). C) The unusual order of the domains of 9th-11th modules of the oxazolomycin pathway made deciphering that part of the pathway difficult. From their signatures, each of the domains in these modules reveal they are mixed with one another. Even so, the ACPs specifically collaborate with the domains of the module to which they belong. Other unusual features are present in the thirteenth module: a DH+ER+KR tridomain that possesses cis-AT architecture, an ACP that clades with cis-AT ACPs, and a docking interaction mediated by catalytically inactive O-methyltransferase (OMT0) and ACP0 domains. Icons and arrowheads are explained in Figure 1.
The current biosynthetic model for the basiliskamide pathway contains several problems (Figure 5a)6, 36. Through elucidating the module types of this assembly line, a correct ordering of its polypeptides can be established (BasG, BasF, BasE, BasD, and BasC). The ACP in BasG is from a v module [GCN5-related acyltransferase (GNAT)-containing] and is thus the first in the assembly line. The ACP in BasF is from a j module, and the DH+KRIII missing from BasF is located at the C-terminal end of BasG. The first ACP of BasE is from a b module, and the KRIII missing from BasE is located at the C-terminal end of BasF. The tandem ACPs in BasD belong to the second module of a type B dehydrating bimodule of which the first module is located at the C-terminal end of BasE. The C domain encoded by BasC apparently substitutes for the second KS of type B dehydrating bimodules. Interestingly, this C domain and a TE that makes an equivalent substitution in the macrolactin assembly line both gatekeep for a α/β-cis, γ/δ-trans diene in contrast to the α/β-trans, γ/δ-cis diene normally selected by type B dehydrating bimodules (the dienes may be interconverted by DH)24.
As the first part of the myxovirescin assembly line generates an unusual intermediate, the order and function of its first polypeptides (TaI, TaL, and Ta-1) had been difficult to assign (Figure 5b)9, 37. However, by determining the module type to which its ACPs belong (the last ACP of TaI and the first KS of TaL collaborate in a d module and that the last ACP of TaL and the first C domain of Ta-1 collaborate in a u module) the polypeptides of the assembly line can be ordered TaI, TaL, Ta-1. The first module, which contains an MT, a GNAT, an ACP, and a KS, likely generates a propionyl group similar to the saxitoxin, enacyloxin, and YM-47522 (related to basiliskamide) pathways38. The second module likely reduces the β-ketone with a B-type KR to generate a D-β-hydroxy group and oxidizes the α-methylene with a trans-Ox to generate a D-α-hydroxy group (d modules are only known to generate D-α, D-β-dihydroxy and L-α, L-β-dihydroxy moieties). The third module may operate similar to Cal13 to generate an α-keto group with its DH39 before its KR performs an α-ketoreduction similar to PksKR3 to install an L-α-hydroxy group22. The KS0 of the third module serves as an adapter, passing the acyl chain to an ACP that functions with its cognate C domain in the fourth module. Another correction to the myxovirescin biosynthetic model is in the seventh module, which possesses the signatures of a d module and generates a D-α, D-β-dihydroxy moiety.
The oxazolomycin assembly line contains several unusual features that had made proposing its biosynthetic model challenging (Figure 5c)6, 40. In OzmH, the 9th–11th modules mix with one another. All of the domains of a type A dehydrating bimodule that comprises the 9th and 10th modules (k & m modules: KRIV+ACP+KS0IV+DH+ACP+KSI,) and all of the domains of the 11th module (b module: KRIII+MT+ACP+KSIII) possess their expected signatures. An unexpected activity is catalyzed by DH, which apparently helps to generate an α/β-trans double bond instead of an anticipated α/β-cis double bond (a leucine substitutes for the conserved active site aspartate). Even with the domains of three modules mixed together, the ACPs only collaborate with their cognate enzymes. The thirteenth module, which contains the DH+ER+KR architecture of cis-AT modules and is split between polypeptides, apparently uses inactive OMT0 and ACP0 domains to noncovalently assemble.
Predicting the stereochemistry of the moiety a module adds is not always possible through a determination of its module type alone. Some module types (e.g., b, d, e, f) include both modules containing A-type KRs that generate L-β-hydroxyl groups and modules containing B-type KRs that generate D-β-hydroxyl groups. In modules with an active DH (e & f) this results in the formation of α/β-trans double bonds from D-β-hydroxyacyl substrates and α/β-cis double bonds from L-β-hydroxyacyl substrates (e.g., Dsz5, Dsz6, Dsz8, Mgs2, Ozm4, and Sor24, which represent a significant fraction of the identified e modules, generate cis-double bonds). KSs of b and d modules are apparently quite tolerant of the stereochemical orientations of α- and β-substituents in the acyl chains they accept, while the KSIs of e and f modules apparently gatekeep for planar geometry at the α- and β-carbons and are able to select for both α/β-trans double bonds and α/β-cis double bonds. The absence or presence of a signature aspartate in the KR usually indicates whether it is A-type or B-type, respectively, although the A-type KRIVs of d modules are an exception to this22, 41.
Insights into Module Function
ACPs that belong to different module types within the same family often possess similar signatures (Figure 4). In Family III, the ACPs from the j module type collaborate with a DH, KR, MT, and an ER. A significant interface is likely formed by ACP with each of these processing enzymes through helices II and III. Even though the ACPs of the b and x module types that are also in Family III do not interact with all of these processing enzymes, their signatures are very similar to those of the j module type. Thus, b and x modules, which likely evolved from j modules, still rely on interactions between their processing enzymes and ACP surfaces generated by the signature residues of Family III.
While ACPs generally co-evolve with the KS of their module type, they can function with other ETR enzymes that have naturally replaced the KS. If the KS that was substituted was not a gatekeeper the replacement does not impact the structure of the intermediate passed to the next module. An example of this innocuous substitution is provided by the bacillaene assembly lines of Bacillus subtilis and Bacillus amyloliquefaciens. After the a-type 11th module of the B. subtilis assembly line has added an L-β-hydroxy unit to the chain, its KS0 transfers the acyl chain to the ACP of the u-type 12th module, which interfaces with a C domain. In the B. amyloliquefaciens assembly line, the 11th module contains a C domain equivalent to that of the 12th module of the B. subtilis assembly line. Presumably, both the KS0 of an ancestral 11th module as well as the ACP of an ancestral 12th module were deleted, and the ACP of the a module was able to collaborate with the C domain of the u module. When gatekeeping KSs are replaced, the new ETR enzyme must select between interconverting acyl chains for the hybrid module to generate a single intermediate. Only four examples of this are present in the 33 analyzed assembly lines (the TE in Dif15 may select for an β-exomethylene, the C domain in Sgv3 may select for an α/β-olefin, the TE in Mln12 may select for an α/β-cis, γ/δ-trans diene, and the C domain of Bas6 may select for an α/β-cis, γ/δ-trans diene).
Evolutionary Relationship between trans-AT and cis-AT Assembly Lines
trans-AT and cis-AT assembly lines have been proposed to have arisen through convergent evolution9, 42; however, the compatibility of trans-AT and cis-AT machinery is suggestive of divergent evolution. Several hybrid assembly lines composed of both trans-AT and cis-AT modules are now known. The chivosazole, enacyloxin, kirromycin, and nosperin assembly lines are primarily comprised of trans-AT modules, the ambruticin, DKxanthene, and pyoluteorin assembly lines are primarily comprised of cis-AT modules, and the neocarzilin assembly line contains an equal mix of trans-AT and cis-AT modules43.
Intriguingly, some modules contain both trans-AT and cis-AT components. The fourteenth module of the kirromycin assembly line appears to be a trans-AT module until closer inspection. The polypeptides KirV (containing the DH+KR+ACP of the fourteenth module) and KirVI (containing the KS of the fourteenth module) noncovalently associate through the same type of docking domains utilized in cis-AT assembly lines44. Its ACP and KS also clade with the ACPs and KSs of cis-AT assembly lines (Figures 2 & 3). The thirteenth module of the oxazolomycin trans-AT assembly line has characteristics of a cis-AT module: its DH, KR, and ER domains are organized as in cis-AT modules, and its ACP contains the “DSLTAVELRN” sequence motif characteristic of ACPs from cis-AT modules (Figure 4). The second and seventh modules of the enacyloxin trans-AT assembly line appear to be cis-AT modules except for their docking domains, which are more like the four-helix bundle docking domains utilized by trans-AT assembly lines23.
cis-AT modules may have evolved through an ancestral trans-AT module. In comparison to the ACPs and KSs from trans-AT modules, the ACPs and KSs of cis-AT modules are much less diverse and cluster tightly with one another in the ACP and KS cladograms (Figures 2 & 3). As the ACPs of cis-AT assembly lines have a “DSLTAVELRN” sequence motif, which is most similar to the signature motif of ACPs from j modules (DH+KRIII+MT+ACP+ER+KSIII), perhaps cis-AT modules arose from such a module (the thirteenth module of the oxazolomycin assembly line, in which the ER domain is positioned between the structural and catalytic subdomains of KR, as in most cis-AT modules, is a good candidate) (Figure 5)45. The “DSLTAVELRN” sequence motif of cis-AT ACPs seems to be important for ACPs that cooperate with an embedded DH, KR, and ER since it is highly conserved in modules that possess these three enzymes and poorly conserved in modules lacking them. That the ACP of the ninth module of the FD-891 cis-AT assembly line possesses the “DSLTAVELRN” motif even though ER is positioned within DH instead of KR indicates that the location of ER within the module does not significantly affect how it binds with ACP46.
Nomenclature
Since transitioning from the traditional module boundaries to the updated module boundaries will likely be a slow process, authors should make the numbering system they are employing evident. The new module boundaries do not change the numbering of KS or C domains (Figure 1). The first module of an assembly line includes the KS or C domain that performs the first chain extension and all the embedded domains upstream of it. All of the ensuing KS, KS0, and C domains are consecutively numbered. The chain releasing enzyme also receives a module number. The numbering of all embedded processing enzymes and ACPs increases by one relative to the traditional numbering. The analysis presented here was aided by assigning letters to module types and numbers to ACP families as well as KS and KR clades; these labels are a temporary convenience and will change as more is learned about the modules of trans-AT assembly lines.
The updated PKS module boundaries affect the boundaries of NRPS modules within trans-AT assembly lines. The NRPS modules of the albicidin assembly line, traditionally defined as C+A+ACP, become A+ACP+C (Ref. 6). As many C domains are known to act as gatekeepers that collaborate with upstream processing enzymes (e.g., E domains), these boundaries make sense7; however, the NRPS community will need to judge whether the NRPS module should be redefined.
Conclusion
Many attempts to Lego-ize enzymatic assembly lines have been made through the years47–48. However, re-engineered assembly lines have traditionally suffered from inactivity or low activity. The newly defined module boundaries afford exciting new opportunities. By keeping together the processing enzymes, ACP, and KS that have co-evolved, it may be possible to engineer assembly lines with activities comparable to those naturally observed. Such evolution-guided engineering could ultimately lead to the programmed synthesis of designer molecules and medicines.
Supplementary Material
Acknowledgments
We thank Christopher N. Boddy and Jessica L. Meinke for their helpful input. This work is supported by grants from NIH (GM106112) and the Welch Foundation (F-1712).
Footnotes
Conflict of Interest
The authors have no conflict of interest to declare.
References
- 1.Mercer AC, Burkart MD. The Ubiquitous Carrier Protein--a Window to Metabolite Biosynthesis. Nat Prod Rep. 2007;24:750–773. doi: 10.1039/b603921a. [DOI] [PubMed] [Google Scholar]
- 2.Crosby J, Crump MP. The Structural Role of the Carrier Protein--Active Controller or Passive Carrier. Nat Prod Rep. 2012;29:1111–1137. doi: 10.1039/c2np20062g. [DOI] [PubMed] [Google Scholar]
- 3.Byers DM, Gong H. Acyl Carrier Protein: Structure-Function Relationships in a Conserved Multifunctional Protein Family. Biochem Cell Biol. 2007;85:649–662. doi: 10.1139/o07-109. [DOI] [PubMed] [Google Scholar]
- 4.Chan DI, Vogel HJ. Current Understanding of Fatty Acid Biosynthesis and the Acyl Carrier Protein. Biochem J. 2010;430:1–19. doi: 10.1042/BJ20100462. [DOI] [PubMed] [Google Scholar]
- 5.Keatinge-Clay AT. Polyketide Synthase Modules Redefined. Angew Chem Int Ed Engl. 2017;56:4658–4660. doi: 10.1002/anie.201701281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helfrich EJ, Piel J. Biosynthesis of Polyketides by Trans-at Polyketide Synthases. Nat Prod Rep. 2016;33:231–316. doi: 10.1039/c5np00125k. [DOI] [PubMed] [Google Scholar]
- 7.Sussmuth RD, Mainz A. Nonribosomal Peptide Synthesis-Principles and Prospects. Angew Chem Int Ed Engl. 2017;56:3770–3821. doi: 10.1002/anie.201609079. [DOI] [PubMed] [Google Scholar]
- 8.Keatinge-Clay AT. The Structures of Type I Polyketide Synthases. Nat Prod Rep. 2012;29:1050–1073. doi: 10.1039/c2np20019h. [DOI] [PubMed] [Google Scholar]
- 9.Piel J. Biosynthesis of Polyketides by Trans-at Polyketide Synthases. Nat Prod Rep. 2010;27:996–1047. doi: 10.1039/b816430b. [DOI] [PubMed] [Google Scholar]
- 10.Zhang L, Hashimoto T, Qin B, Hashimoto J, Kozone I, Kawahara T, Okada M, Awakawa T, Ito T, Asakawa Y, Ueki M, Takahashi S, Osada H, Wakimoto T, Ikeda H, Shin-Ya K, Abe I. Characterization of Giant Modular Pkss Provides Insight into Genetic Mechanism for Structural Diversification of Aminopolyol Polyketides. Angew Chem Int Ed Engl. 2017;56:1740–1745. doi: 10.1002/anie.201611371. [DOI] [PubMed] [Google Scholar]
- 11.Piel J. A Polyketide Synthase-Peptide Synthetase Gene Cluster from an Uncultured Bacterial Symbiont of Paederus Beetles. Proc Natl Acad Sci U S A. 2002;99:14002–14007. doi: 10.1073/pnas.222481399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng YQ, Tang GL, Shen B. Type I Polyketide Synthase Requiring a Discrete Acyltransferase for Polyketide Biosynthesis. Proc Natl Acad Sci U S A. 2003;100:3149–3154. doi: 10.1073/pnas.0537286100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen T, Ishida K, Jenke-Kodama H, Dittmann E, Gurgui C, Hochmuth T, Taudien S, Platzer M, Hertweck C, Piel J. Exploiting the Mosaic Structure of Trans-Acyltransferase Polyketide Synthases for Natural Product Discovery and Pathway Dissection. Nat Biotechnol. 2008;26:225–233. doi: 10.1038/nbt1379. [DOI] [PubMed] [Google Scholar]
- 14.Haines AS, Dong X, Song Z, Farmer R, Williams C, Hothersall J, Ploskon E, Wattana-Amorn P, Stephens ER, Yamada E, Gurney R, Takebayashi Y, Masschelein J, Cox RJ, Lavigne R, Willis CL, Simpson TJ, Crosby J, Winn PJ, Thomas CM, Crump MP. A Conserved Motif Flags Acyl Carrier Proteins for Beta-Branching in Polyketide Synthesis. Nat Chem Biol. 2013;9:685–692. doi: 10.1038/nchembio.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Medema MH, Kottmann R, Yilmaz P, Cummings M, Biggins JB, Blin K, de Bruijn I, Chooi YH, Claesen J, Coates RC, Cruz-Morales P, Duddela S, Dusterhus S, Edwards DJ, Fewer DP, Garg N, Geiger C, Gomez-Escribano JP, Greule A, Hadjithomas M, Haines AS, Helfrich EJ, Hillwig ML, Ishida K, Jones AC, Jones CS, Jungmann K, Kegler C, Kim HU, Kotter P, Krug D, Masschelein J, Melnik AV, Mantovani SM, Monroe EA, Moore M, Moss N, Nutzmann HW, Pan G, Pati A, Petras D, Reen FJ, Rosconi F, Rui Z, Tian Z, Tobias NJ, Tsunematsu Y, Wiemann P, Wyckoff E, Yan X, Yim G, Yu F, Xie Y, Aigle B, Apel AK, Balibar CJ, Balskus EP, Barona-Gomez F, Bechthold A, Bode HB, Borriss R, Brady SF, Brakhage AA, Caffrey P, Cheng YQ, Clardy J, Cox RJ, De Mot R, Donadio S, Donia MS, van der Donk WA, Dorrestein PC, Doyle S, Driessen AJ, Ehling-Schulz M, Entian KD, Fischbach MA, Gerwick L, Gerwick WH, Gross H, Gust B, Hertweck C, Hofte M, Jensen SE, Ju J, Katz L, Kaysser L, Klassen JL, Keller NP, Kormanec J, Kuipers OP, Kuzuyama T, Kyrpides NC, Kwon HJ, Lautru S, Lavigne R, Lee CY, Linquan B, Liu X, Liu W, Luzhetskyy A, Mahmud T, Mast Y, Mendez C, Metsa-Ketela M, Micklefield J, Mitchell DA, Moore BS, Moreira LM, Muller R, Neilan BA, Nett M, Nielsen J, O’Gara F, Oikawa H, Osbourn A, Osburne MS, Ostash B, Payne SM, Pernodet JL, Petricek M, Piel J, Ploux O, Raaijmakers JM, Salas JA, Schmitt EK, Scott B, Seipke RF, Shen B, Sherman DH, Sivonen K, Smanski MJ, Sosio M, Stegmann E, Sussmuth RD, Tahlan K, Thomas CM, Tang Y, Truman AW, Viaud M, Walton JD, Walsh CT, Weber T, van Wezel GP, Wilkinson B, Willey JM, Wohlleben W, Wright GD, Ziemert N, Zhang C, Zotchev SB, Breitling R, Takano E, Glockner FO. Minimum Information About a Biosynthetic Gene Cluster. Nat Chem Biol. 2015;11:625–631. doi: 10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edgar RC. Muscle: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic acids research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stamatakis A. Raxml Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le SQ, Gascuel O. An Improved General Amino Acid Replacement Matrix. Molecular biology and evolution. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 19.Huson DH, Scornavacca C. Dendroscope 3: An Interactive Tool for Rooted Phylogenetic Trees and Networks. Systematic biology. 2012;61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- 20.Lohman JR, Ma M, Osipiuk J, Nocek B, Kim Y, Chang C, Cuff M, Mack J, Bigelow L, Li H, Endres M, Babnigg G, Joachimiak A, Phillips GN, Jr, Shen B. Structural and Evolutionary Relationships of “at-Less” Type I Polyketide Synthase Ketosynthases. Proc Natl Acad Sci U S A. 2015;112:12693–12698. doi: 10.1073/pnas.1515460112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gay DC, Gay G, Axelrod AJ, Jenner M, Kohlhaas C, Kampa A, Oldham NJ, Piel J, Keatinge-Clay AT. A Close Look at a Ketosynthase from a Trans-Acyltransferase Modular Polyketide Synthase. Structure. 2014;22:444–451. doi: 10.1016/j.str.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piasecki SK, Zheng J, Axelrod AJ, Detelich ME, Keatinge-Clay AT. Structural and Functional Studies of a Trans-Acyltransferase Polyketide Assembly Line Enzyme That Catalyzes Stereoselective Alpha- and Beta-Ketoreduction. Proteins. 2014;82:2067–2077. doi: 10.1002/prot.24561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeng J, Wagner DT, Zhang Z, Moretto L, Addison JD, Keatinge-Clay AT. Portability and Structure of the Four-Helix Bundle Docking Domains of Trans-Acyltransferase Modular Polyketide Synthases. ACS Chem Biol. 2016;11:2466–2474. doi: 10.1021/acschembio.6b00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wagner DT, Zeng J, Bailey CB, Gay DC, Yuan F, Manion HR, Keatinge-Clay AT. Structural and Functional Trends in Dehydrating Bimodules from Trans-Acyltransferase Polyketide Synthases. Structure. 2017;25:1045–1055. e1042. doi: 10.1016/j.str.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gay DC, Spear PJ, Keatinge-Clay AT. A Double-Hotdog with a New Trick: Structure and Mechanism of the Trans-Acyltransferase Polyketide Synthase Enoyl-Isomerase. ACS Chem Biol. 2014;9:2374–2381. doi: 10.1021/cb500459b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crooks GE, Hon G, Chandonia JM, Brenner SE. Weblogo: A Sequence Logo Generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, Liu N, Cacho RA, Gong Z, Liu Z, Qin W, Tang C, Tang Y, Zhou J. Structural Basis of Nonribosomal Peptide Macrocyclization in Fungi. Nat Chem Biol. 2016;12:1001–1003. doi: 10.1038/nchembio.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drake EJ, Miller BR, Shi C, Tarrasch JT, Sundlov JA, Allen CL, Skiniotis G, Aldrich CC, Gulick AM. Structures of Two Distinct Conformations of Holo-Non-Ribosomal Peptide Synthetases. Nature. 2016;529:235–238. doi: 10.1038/nature16163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell CA, Shi C, Aldrich CC, Gulick AM. Structure of Pa1221, a Nonribosomal Peptide Synthetase Containing Adenylation and Peptidyl Carrier Protein Domains. Biochemistry. 2012;51:3252–3263. doi: 10.1021/bi300112e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen WH, Li K, Guntaka NS, Bruner SD. Interdomain and Intermodule Organization in Epimerization Domain Containing Nonribosomal Peptide Synthetases. ACS Chem Biol. 2016;11:2293–2303. doi: 10.1021/acschembio.6b00332. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y, Zheng T, Bruner SD. Structural Basis for Phosphopantetheinyl Carrier Domain Interactions in the Terminal Module of Nonribosomal Peptide Synthetases. Chem Biol. 2011;18:1482–1488. doi: 10.1016/j.chembiol.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haslinger K, Brieke C, Uhlmann S, Sieverling L, Sussmuth RD, Cryle MJ. The Structure of a Transient Complex of a Nonribosomal Peptide Synthetase and a Cytochrome P450 Monooxygenase. Angew Chem Int Ed Engl. 2014;53:8518–8522. doi: 10.1002/anie.201404977. [DOI] [PubMed] [Google Scholar]
- 33.Miyanaga A, Iwasawa S, Shinohara Y, Kudo F, Eguchi T. Structure-Based Analysis of the Molecular Interactions between Acyltransferase and Acyl Carrier Protein in Vicenistatin Biosynthesis. Proc Natl Acad Sci U S A. 2016;113:1802–1807. doi: 10.1073/pnas.1520042113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rahman AS, Hothersall J, Crosby J, Simpson TJ, Thomas CM. Tandemly Duplicated Acyl Carrier Proteins, Which Increase Polyketide Antibiotic Production, Can Apparently Function Either in Parallel or in Series. Journal of Biological Chemistry. 2005;280:6399–6408. doi: 10.1074/jbc.M409814200. [DOI] [PubMed] [Google Scholar]
- 35.Ma M, Lohman JR, Liu T, Shen B. C-S Bond Cleavage by a Polyketide Synthase Domain. Proc Natl Acad Sci U S A. 2015;112:10359–10364. doi: 10.1073/pnas.1508437112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Theodore CM, Stamps BW, King JB, Price LS, Powell DR, Stevenson BS, Cichewicz RH. Genomic and Metabolomic Insights into the Natural Product Biosynthetic Diversity of a Feral-Hog-Associated Brevibacillus Laterosporus Strain. PLoS One. 2014;9:e90124. doi: 10.1371/journal.pone.0090124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simunovic V, Zapp J, Rachid S, Krug D, Meiser P, Muller R. Myxovirescin a Biosynthesis Is Directed by Hybrid Polyketide Synthases/Nonribosomal Peptide Synthetase, 3-Hydroxy-3-Methylglutaryl-Coa Synthases, and Trans-Acting Acyltransferases. Chembiochem. 2006;7:1206–1220. doi: 10.1002/cbic.200600075. [DOI] [PubMed] [Google Scholar]
- 38.Kellmann R, Mihali TK, Jeon YJ, Pickford R, Pomati F, Neilan BA. Biosynthetic Intermediate Analysis and Functional Homology Reveal a Saxitoxin Gene Cluster in Cyanobacteria. Appl Environ Microbiol. 2008;74:4044–4053. doi: 10.1128/AEM.00353-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wakimoto T, Egami Y, Nakashima Y, Wakimoto Y, Mori T, Awakawa T, Ito T, Kenmoku H, Asakawa Y, Piel J, Abe I. Calyculin Biogenesis from a Pyrophosphate Protoxin Produced by a Sponge Symbiont. Nat Chem Biol. 2014;10:648–655. doi: 10.1038/nchembio.1573. [DOI] [PubMed] [Google Scholar]
- 40.Zhao C, Ju J, Christenson SD, Smith WC, Song D, Zhou X, Shen B, Deng Z. Utilization of the Methoxymalonyl-Acyl Carrier Protein Biosynthesis Locus for Cloning the Oxazolomycin Biosynthetic Gene Cluster from Streptomyces Albus Ja3453. J Bacteriol. 2006;188:4142–4147. doi: 10.1128/JB.00173-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keatinge-Clay AT. Stereocontrol within Polyketide Assembly Lines. Nat Prod Rep. 2016;33:141–149. doi: 10.1039/c5np00092k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jenke-Kodama H, Sandmann A, Muller R, Dittmann E. Evolutionary Implications of Bacterial Polyketide Synthases. Mol Biol Evol. 2005;22:2027–2039. doi: 10.1093/molbev/msi193. [DOI] [PubMed] [Google Scholar]
- 43.Keatinge-Clay AT. The Uncommon Enzymology of Cis-Acyltransferase Assembly Lines. Chem Rev. 2017;117:5334–5366. doi: 10.1021/acs.chemrev.6b00683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weissman KJ. The Structural Basis for Docking in Modular Polyketide Biosynthesis. Chembiochem. 2006;7:485–494. doi: 10.1002/cbic.200500435. [DOI] [PubMed] [Google Scholar]
- 45.Zheng JT, Gay DC, Demeler B, White MA, Keatinge-Clay AT. Divergence of Multimodular Polyketide Synthases Revealed by a Didomain Structure. Nature Chemical Biology. 2012;8:615–621. doi: 10.1038/nchembio.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kudo F, Motegi A, Mizoue K, Eguchi T. Cloning and Characterization of the Biosynthetic Gene Cluster of 16-Membered Macrolide Antibiotic Fd-891: Involvement of a Dual Functional Cytochrome P450 Monooxygenase Catalyzing Epoxidation and Hydroxylation. Chembiochem. 2010;11:1574–1582. doi: 10.1002/cbic.201000214. [DOI] [PubMed] [Google Scholar]
- 47.Menzella HG, Reid R, Carney JR, Chandran SS, Reisinger SJ, Patel KG, Hopwood DA, Santi DV. Combinatorial Polyketide Biosynthesis by De Novo Design and Rearrangement of Modular Polyketide Synthase Genes. Nat Biotechnol. 2005;23:1171–1176. doi: 10.1038/nbt1128. [DOI] [PubMed] [Google Scholar]
- 48.Sherman DH. The Lego-Ization of Polyketide Biosynthesis. Nat Biotechnol. 2005;23:1083–1084. doi: 10.1038/nbt0905-1083. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.