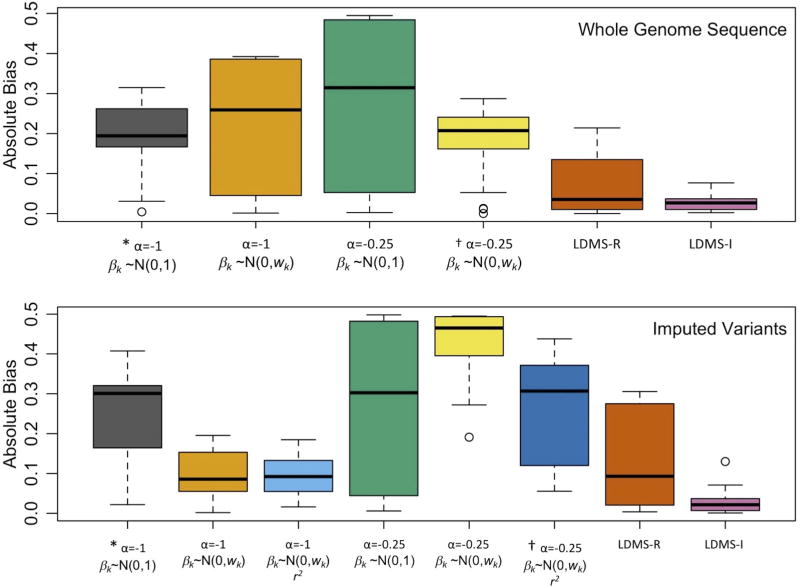
Figure 5.
Boxplots of the absolute bias of heritability estimates across all simulated phenotypes from Supplementary Figures 24 & 26 using WGS data to estimate GRMs (top), and from Figures 3–4 using imputed variants to estimate the GRMs (bottom). X axis indicates the parameters for the estimation model, including the MAF scaling factor, α, and the assumed effect size distribution, βk, specified in the GRM and whether imputation scores (r2) were used in the GRM estimation. All used a single GRM except for LD- & MAF-stratified GREML (LDMS), which used 16 GRMs (α=−1) stratified by MAF and either regional (-R) or individual SNP (-I) LD score. * Typical GREML-SC parameters. † Typical LDAK-SC parameters. Boxplots show the median and interquartile, with whiskers extending 1.5 times the quartiles and more extreme points shown for N=22 (WGS) and 26 (imputed) mean estimates of heritability.