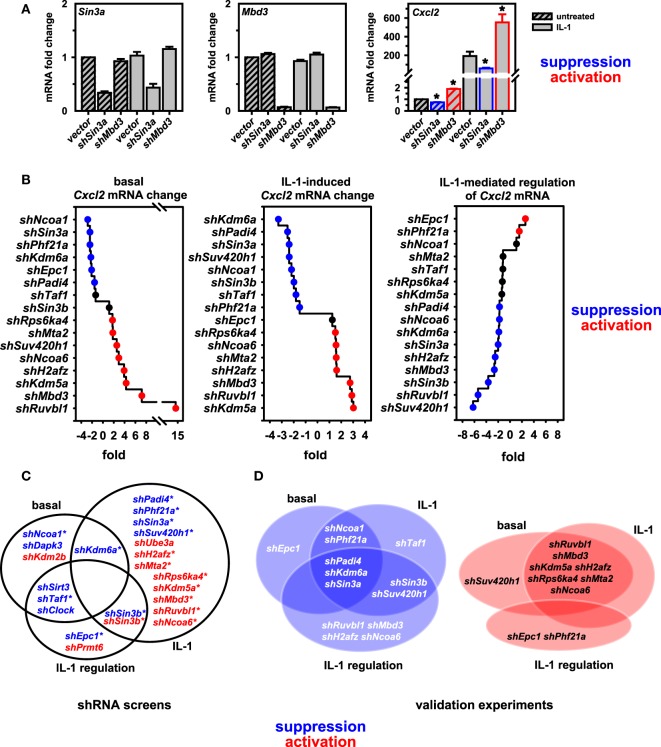
Figure 3.
Validation of screen results further defines coactivators and corepressors of Cxcl2. (A) Murine embryonic fibroblast (Mef) cells were transfected with empty pLKO.1 or with pooled shRNAs directed against Sin3a or Mbd3. After 48 h of selection in puromycin, cells were treated with IL-1 for 1 h or were left untreated. Then, total RNA was isolated and mRNA expression of Cxcl2, Sin3a, Mbd3 and Ube2l3 was determined by conventional RT-qPCR. Expression values of Cxcl2, Sin3a, or Mbd3 were normalized for expression of Ube2l3. The graphs show mean relative expression values ± SEM relative to the vector control from two independent series of experiments performed in duplicates. Asterisks indicate significant changes (p < 0.05) derived from Mann-Whitney Rank Sum t-tests. (B) The same RT-qPCR approach was used to determine the effects of 16 further knockdowns of nuclear cofactors on basal and IL-1-inducible Cxcl2 levels as described in detail in the legend of Figure S5 in Supplementary Material. The line graphs show the ranked mean fold changes of normalized Cxcl2 mRNA expression separately analyzed for basal and IL-1-induced conditions. Additionally, the effects on IL-1-mediated regulation (as defined in Figure S3 in Supplementary Material) are depicted. (C) Summarizing venn diagrams indicating the overlapping effects on Cxcl2 mRNA expression of shRNAs directed against 22 nuclear cofactors as identified by shRNA screens I and II. Asterisks indicate the 16 factors that were chosen for validation. (D) Overlap of functions of the shRNAs as found by conventional RT-qPCR. (A–D) Red or blue colors indicate at least 1.5-fold differences between shRNA versus empty vector transfections.