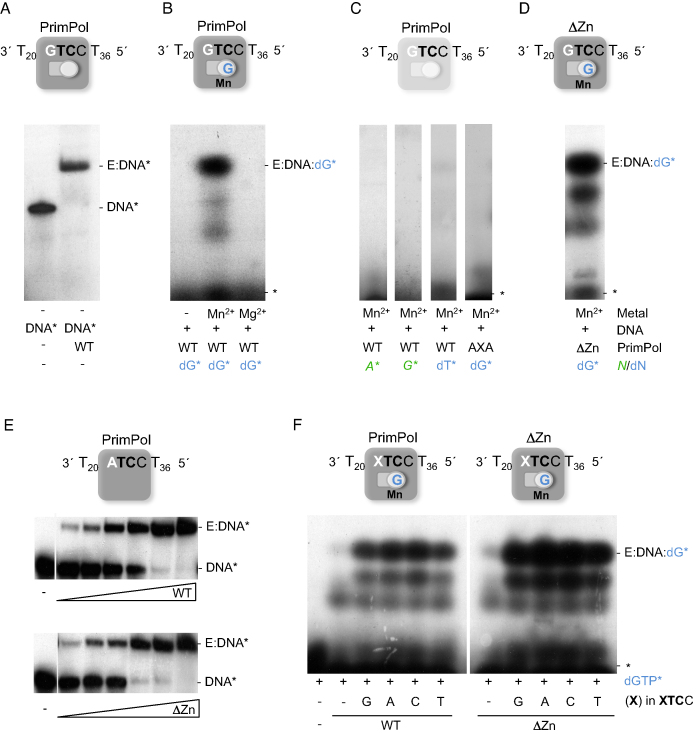
Figure 2.
PrimPol binds the GTCC oligonucleotide and the 3′-site nucleotide dGTP to establish the pre-ternary complex. (A) Binary complex (E:DNA*) of PrimPol (40 nM) with [γ-32P]-labeled oligonucleotide GTCC (1 nM), as detected by EMSA. The scheme above shows the ssDNA and PrimPol, with both 5′-nucleotide (primer; square) and 3′-nucleotide (incoming; circle) binding sites empty. (B) Formation of a pre-ternary complex (E:DNA:dG*) when providing PrimPol (0.5 μM), non-labeled ssDNA GTCC (1 μM), MnCl2 (1 mM), and labeled 3′-nucleotide [α-32P]dGTP (16 nM), which occupies its position as shown in the scheme above. (C) A pre-ternary complex was not detected by EMSA either with [γ-32P]ATP, [γ-32P]GTP, a non-complementary 3′-deoxynucleotide [α32P]-dTTP (16 nM), or with a PrimPol mutant lacking two catalytic metal ligands (AxA). (D) Pre-ternary complex formed with ΔZnFD mutant (ΔZn) (0.5 μM), ssDNA GTCC (1 μM), [α-32P]dGTP (16 nM), and 1 mM MnCl2. (E) EMSA showing the interaction of PrimPol WT or ΔZnFD mutant (2.5, 5, 10, 20, 40, 80 nM) with [γ-32P]-labeled ATCC oligonucleotide (60-mer; 1 nM) in the absence of metal cofactor. (F) Pre-ternary complex of either PrimPol or ΔZnFD mutant (ΔZn) (0.5 μM) with [α-32P]dGTP (16 nM), 1 mM MnCl2 and different oligonucleotides (XTCC), where the cryptic G has been replaced by each of the three other nucleotides A, C and T (depicted as an X in the scheme; 1 μM). The mobility of free labeled ssDNA (DNA*), binary complex (E:DNA*), free labeled nucleotide (*), or pre-ternary complex (E:DNA:dG*) are indicated in the gels. The autoradiographs shown in this figure are representative of at least 3 independent EMSA experiments.