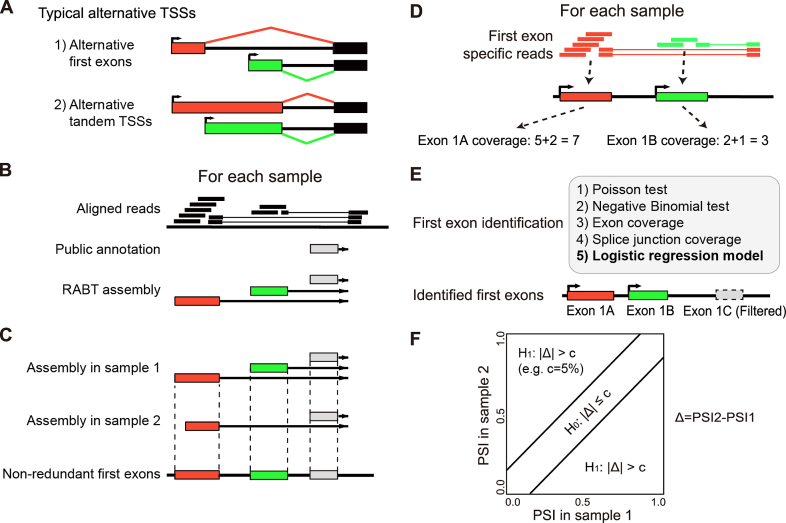
Figure 1.
The SEASTAR pipeline for the computational identification and quantitative analysis of first exons using RNA-seq data alone. (A) Alternative transcription start sites (TSSs) can appear in two forms: alternative first exons (AFEs) and alternative tandem TSSs. (B) The reference guided transcript assembly: the reference annotation based transcript (RABT) assembly method is used to assemble novel transcripts using RNA-seq reads guided by the existing transcriptome annotation. (C) The generation of non-redundant first exons (FEs): transcripts from all samples are merged to generate a non-redundant set of FEs. (D) The quantitation of exon and splice junction coverage: reads mapped to each FE and its downstream splice junction are counted as the coverage for each FE. (E) The identification of bona fide FEs: five methods are designed and compared. The logistic regression model (highlighted in bold) is selected as the method of choice in SEASTAR due to its superior performance. (F) The detection of differential AFE usage: the percent-spliced-in (PSI) value for each AFE in each sample is calculated using the read counts and effective lengths of all AFEs within the gene. The rMATS statistical test is used to determine whether the AFE has significant differential usage between two samples or two groups of samples.