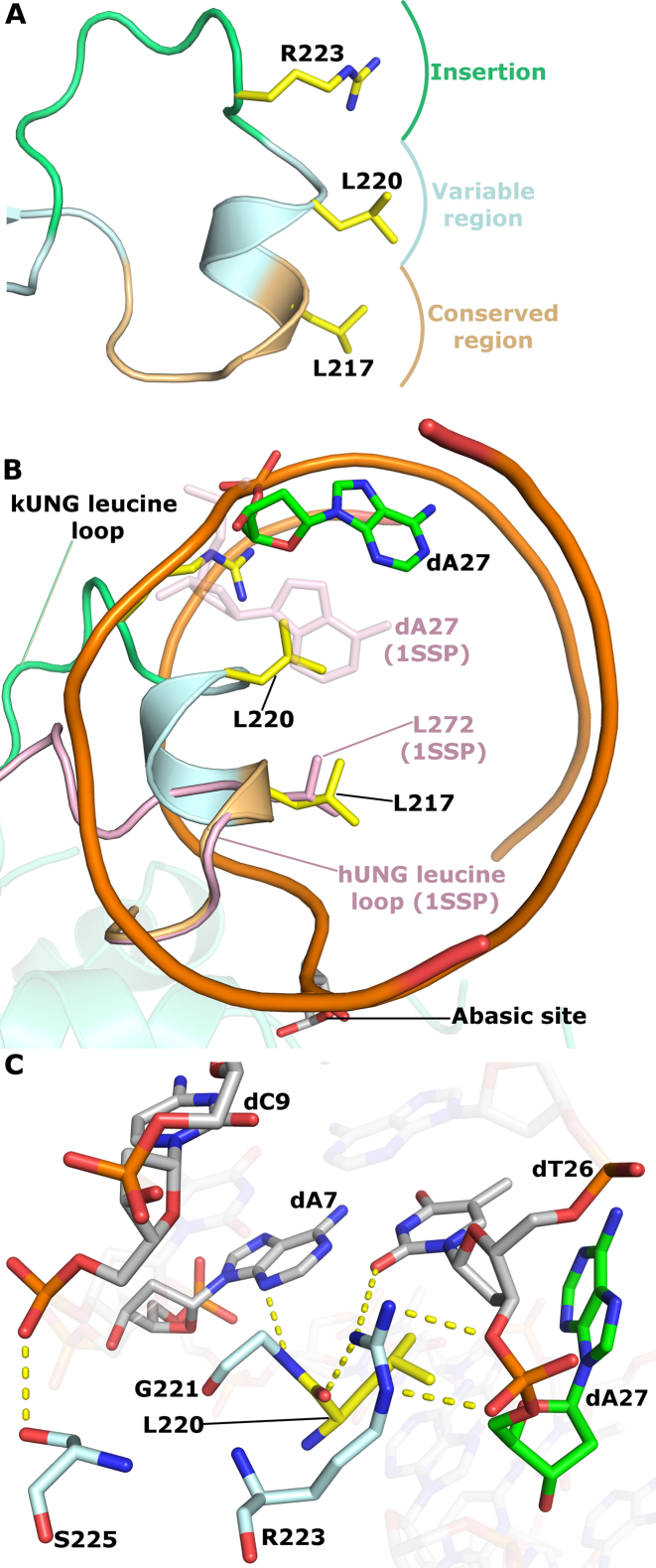
Figure 5.
kUNG leucine loop structure. (A) Schematic of the conserved, variable and extension regions of the kUNG leucine loop. The strictly conserved ‘doorstop’ leucine L217, orphan nucleotide-flipping leucine L220 and key DNA-binding residue R223 sidechains are shown as sticks with yellow carbons. (B) View along the axis of the DNA double helix comparing kUNG and hUNG leucine loop positions, the DNA backbone is shown as an orange ribbon, most nucleotides are omitted for clarity. The kUNG leucine loop is coloured [as in panel A], the hUNG leucine loop, from PDB code: 1SSP (31), is shown in pink cartoon representation with the ‘doorstop’ leucine shown as pink sticks. The orphan dA27 nucleotide in the kUNG–dsDNA structure is shown in sticks with green carbons. The orphan base in 1SSP (faded pink sticks) takes its usual position in the base stack; this position would be precluded by steric hindrance from L220 of kUNG. (C) Stick representation of key protein–DNA interactions between the variable/extension regions of the leucine loop and the DNA minor groove. Protein residues are shown as sticks with pale cyan carbons, L220 is shown with yellow carbons. DNA is coloured [as in panel B]. Hydrogen bonds/electrostatic interactions are shown as yellow dashes. The guanidinium head group of R223 interacts with the phosphate group of dA27 and O2 of the dT26 base, as well as the carbonyl oxygen of L220. Further interactions from the kUNG leucine loop in the minor groove come from S225 and G221.