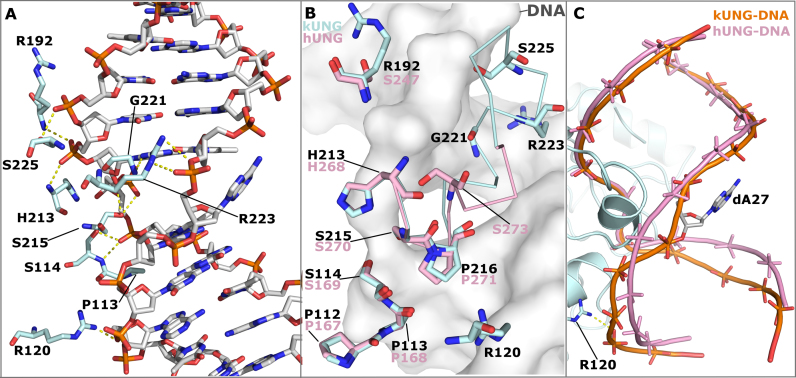
Figure 6.
Protein–DNA contacts in the kUNG–dsDNA complex, see Supplementary Table S1 for details. (A) Stick representation showing DNA-contacting kUNG residues (pale cyan carbons), all other protein residues are omitted for clarity. Hydrogen bonds/electrostatic interactions are shown as yellow dashes. (B) Alignment of kUNG–dsDNA and hUNG–dsDNA (PDB code: 1SSP) complexes displaying structural conservation of the ‘Ser-Pro pinch' residues. kUNG residues are shown as sticks with pale cyan carbons, hUNG residues are shown as sticks with pink carbons. kUNG and hUNG leucine loop backbones are shown as ribbons in pale cyan and pink respectively. DNA from the kUNG–dsDNA complex is shown as a grey surface. Despite excellent overall conservation, the action of S273 in hUNG as a hydrogen bond donor is not mimicked by kUNG. Instead, contributions from G221, R223 and S225 provide ‘pinching’ interactions to compress the DNA backbone. R223 of kUNG provides an additional contact with the DNA backbone not seen in hUNG. (C) Comparison of the global DNA backbone conformation in enzyme–product complexes of kUNG (orange DNA) and hUNG (pink DNA). DNA backbones are shown as ribbon traces between phosphates. 3′ of the uracil (at the top of the image), the DNA backbone position is largely similar. 5′ of the uracil however, the DNA takes up a position closer to kUNG than in hUNG, a position supported by contact between R223 and the phosphate of dA29. There is a less pronounced kink in the DNA by kUNG than is seen in hUNG–dsDNA structures. Consequently, the flipped out orphan base dA27 is presented to the solvent in a more accessible major groove.