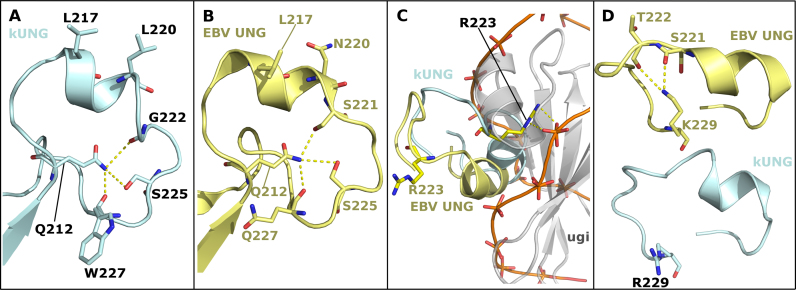
Figure 7.
Comparison of leucine loop structures in the kUNG–dsDNA and EBV UNG–Ugi complexes (PDB code: 2J8X). (A and B) The leucine loops of kUNG (A, pale cyan) and EBV UNG (B, yellow) have similar structures including a rigid conformation of the extension region being centred on hydrogen-bonding with Q212. Residues involved in this hydrogen-bonding network and nucleotide-flipping residues are shown as sticks. Hydrogen bonds/electrostatic interactions are shown as yellow dashes. (C) Relative positions of the kUNG and EBV UNG leucine loops with the two UNGs aligned on conserved catalytic core residues. The kUNG leucine loop (pale cyan) invades the minor groove of the DNA (shown as an orange trace between phosphates). The EBV UNG leucine loop (yellow) is precluded from the same position as the kUNG loop by the bulky nature of Ugi (grey cartoon). R223 forms protein–DNA contacts in kUNG but protrudes into the solvent in the Ugi-bound EBV UNG structure. (D) Relative positions of R/K229. In the EBV UNG–Ugi structure (yellow), the side-chain of K229 is oriented towards the centre of the loop and interacts with T222 and S221. The side-chain of R229 of kUNG (pale cyan) forms no contacts with other atoms. R/K229 may act as a pre-catalytic ‘pinning’ residue to hold the loop away from the DNA binding cleft to allow DNA binding.