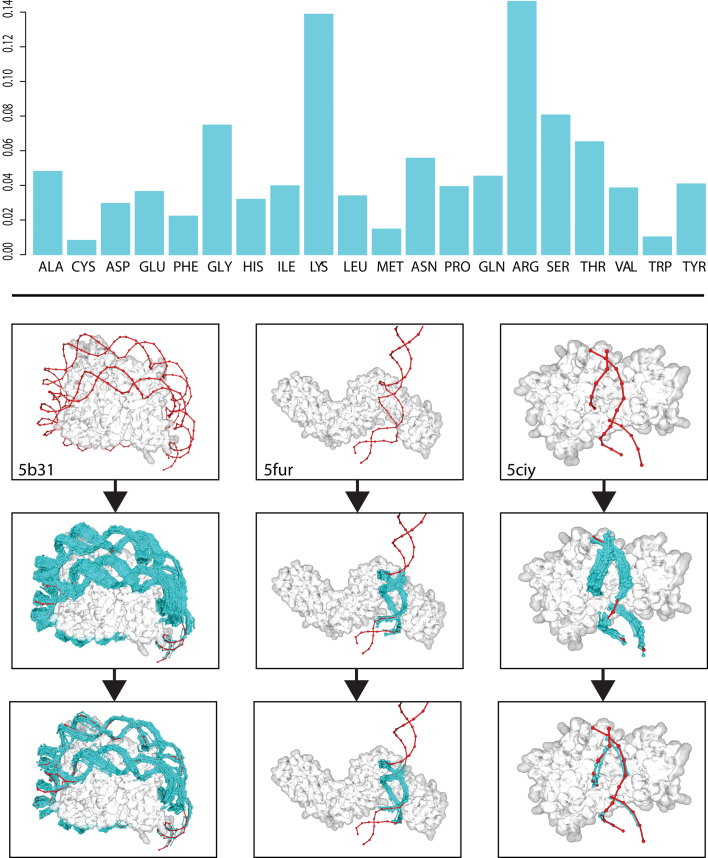
Figure 3.
(Upper) DNA-amino acid binding propensities for all residues against any given nucleotide on the ModelXDB. (Lower) Examples (PDBs: 5b31, 5fur, 5ciy, top panel) of PADA1 predictions: fragment clouds (cyan) are filtered using the PADA1 force field going from a disperse cloud (medium panel) to a refined cluster (bottom panel) containing the most energetically favorable docks. Crystallographic DNA in red.