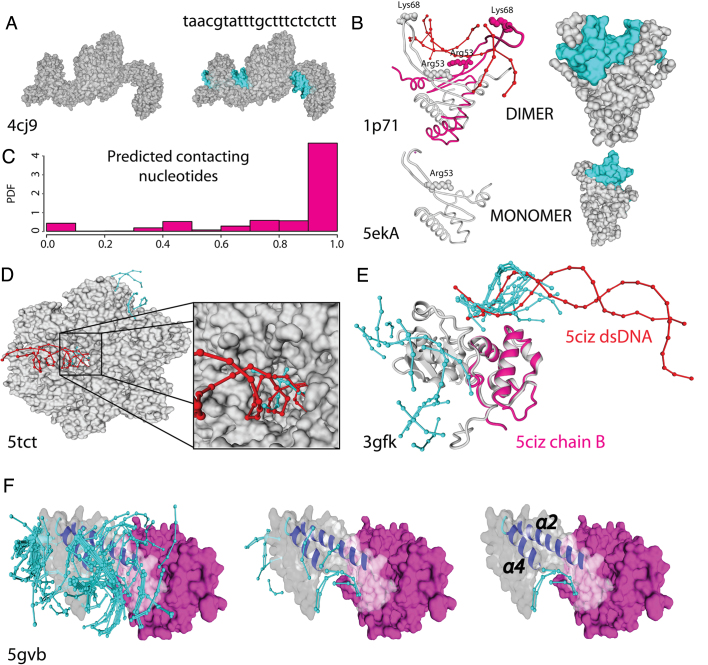
Figure 5.
Accuracy on the docking predictions: In all cases cyan colour is used for docked DNA and red for the crystallographic one. (A) TAL effector (PDB: 4jc9), protein–DNA interface regions and nucleotide sequence specificity are correctly predicted. (B) Left and right: Cartoon and VdW surface style for a dimeric structure with DNA (PDB: 1p71; Upper) and the related protein crystallized as a monomer missing part of the structure (PDB: 5eka; lower). Predicted docked DNA in both structures in cyan. (C) histogram of sequence specificity accuracy over 13 proteins of different PFam family. (D) Humanized yeast ACC carboxyltransferase (PDB: 5tct) binding region (zoomed) was found within the five best energy docks. (E) the DNA-directed RNA polymerase subunit alpha of Bacillus subtilis (PDB: 3gfk, grey) superimposed to the overlapping domain of the E. coli ortholog with low sequence identity (PDB: 5ciz chainB, magenta). (F) The helical bundle of AND-1 human protein (PDB: 5gvb) present a dense cloud of docked fragments (left side) in the binding groove formed by the α2 and α4 helices. Within the six best energy docks for AND-1 human protein we found three docks (right side) placed within the predicted binding region.