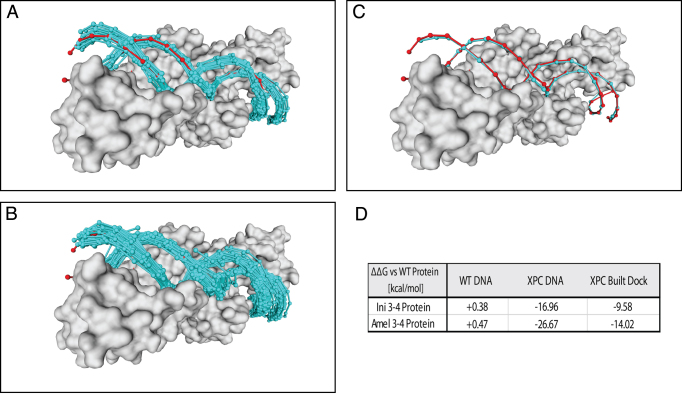
Figure 6.
I-Crel Amel3-4 (PDB: 4aqu) engineered protein: (A) DNA Docked molecules obtained using PADA1 default parameters in blue, XPC DNA in red. (B) Dockings obtained with relaxed parameters (dubiety = 0.5, cb-angle = 8°). (C) The merged DNA fragment using RMSD criteria against XPC DNA superimposed on the crystallographic DNA. (D) Energy variation for both Ini3-4 and Amel3-4 engineered proteins against the different DNAs. The built models show the same specificity tendencies experimentally reported for crystallographic DNAs (WT and XPC), and the PADA1 built dock shows that the full in-silico analysis reproduces the experimental affinity tendencies studied.