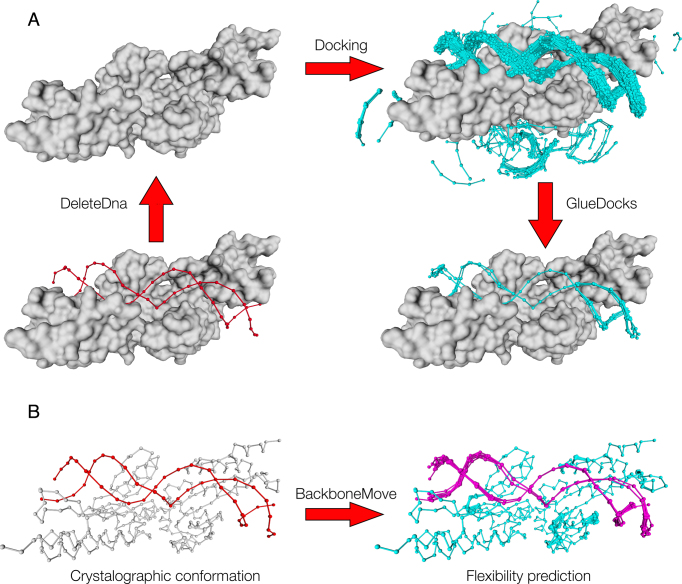
Figure 7.
Modeling of flexibility upon binding. (A) DNA flexibility prediction: We first remove the Crystallographic DNA, then we do DNA Docking and select the fragments that will be used for reconstructing the DNA molecule. Once the fragments are selected we join those that are compatible using (GlueDocks; see Materials and Methods). As can be seen the cleavage site is quite rigid, while the DNA backbone becomes more flexible farther away from it. This flexibility could be used to design protein mutants that will recognize other DNA sequences. (B) Protein flexibility prediction: the BackboneMove command can be used to model protein backbone variability over the high free energy regions generated upon DNA flexibility prediction.