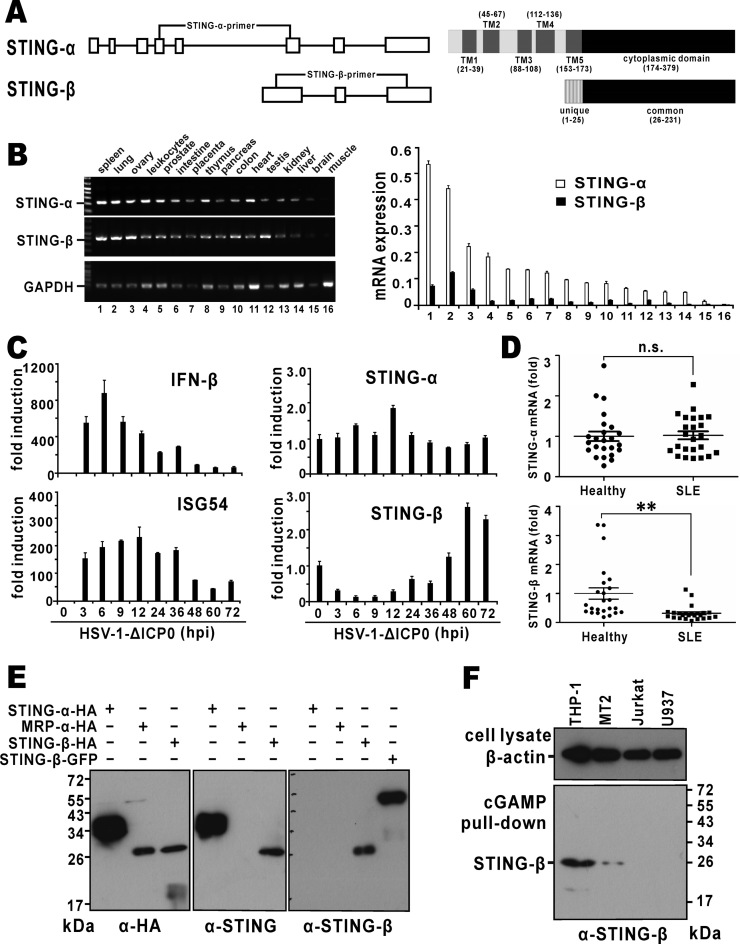
Figure 1.
Expression of STING-β mRNA and protein. (A) Genome and domain structure of STING-β. Boxes represent exons and the lines represent introns. Positions of isoform-specific primers used in RT-PCR are indicated. Also shown are transmembrane (TM) and cytoplasmic domains of STING-α and STING-β. Transcription of STING-β mRNA is driven by an alternative promoter located within intron 5 of STING-α. Compared to exon 6 of STING-α, exon 1 of STING-β contains an extra piece of sequence at the 5′ end, resulting in an additional 25 amino acids at the N-terminus. This unique N-terminal sequence is highlighted. (B) Expression of STING-α and STING-β transcripts in human tissues. Human MTC™ Panel I (Clontech, USA) containing the cDNA templates of various human tissues were used for RT-PCR (left panel) and RT-qPCR (right panel) analysis. The STING-β primers will amplify a fragment of the correct size from STING-β cDNA only, but neither STING-α cDNA nor genomic DNA. Likewise, the STING-α primers are specific to STING-α cDNA. mRNA level was obtained by the comparative Ct method. (C) Expression of STING-β transcript in virus-challenged THP-1 cells. A DNA virus HSV-1-ΔICP0 (5 M.O.I.), in which one major IFN antagonist named ICP0 is deleted, was used to stimulate type I IFN production in THP-1 cells. Samples were harvested at the indicated time points for RNA extraction. Temporal expression profile of STING-β was determined by RT-qPCR. (D) Expression of STING-β transcript in peripheral blood mononuclear cells of SLE patients. cDNA templates were prepared from peripheral blood mononuclear cells of SLE patients (n = 24) and healthy individuals (n = 24). Expression of STING-α and STING-β in theses samples was detected by RT-qPCR. Whereas there was no significant statistical difference (n.s.) in STING-α mRNA expression between the SLE and healthy groups, the levels of STING-β mRNA were significantly lower (∗∗P < 0.01) in SLE samples versus cells from healthy people. Statistical analysis was performed using Student's t test. (E) Detection of recombinant STING-β protein expressed in HEK293T cells by STING-β-specific antibodies. STING-α-HA, STING-β-HA, MRP-HA and STING-β-GFP were overexpressed in HEK293T cells. After 48 h, cells were lysed for western blotting using the indicated antibodies. Rabbit anti-STING-β antiserum specifically recognizes STING-β but not STING-α or MRP. (F) Detection of endogenous STING-β protein by STING-β-specific antibodies. THP-1, MT2, Jurkat and U937 cells were lysed and incubated with cGAMP agarose for 2 h at 4°C. After washing with PBS containing 100 μM ATP for six times, sample loading buffer was added. The samples were boiled for 10 min and then further analyzed by SDS-PAGE and western blotting with anti-STING-β antibodies. Results in each panel are representative of three independent experiments.