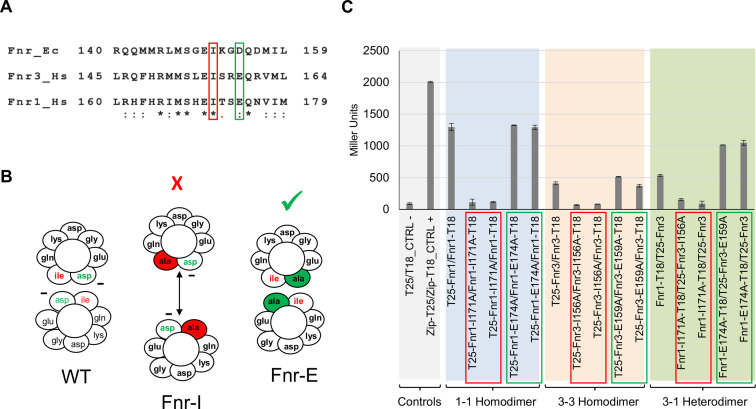
Figure 4.
Interaction between Fnr1 and Fnr3 requires the dimerization helix. (A) Dimerization helix alignment from E. coli Fnr (Fnr_Ec) and H. seropedicae Fnr1 (Fnr1_Hs) and Fnr3 (Fnr3_Hs) proteins. Amino acid residues important for the interaction between subunits are highlighted in red and green. (B) Cross-section of the dimerization interface of wild type Fnr, FnrI151A and FnrD154A. Diagrams are based on the E. coli Fnr sequence (45,46). The charge of Asp154 is partially shielded from the interface by the presence of Ile151. Removal of the large hydrophobic residue at position 151 (Ile → Ala) allows the negative charge of Asp154 to become exposed in the interface and the repulsive interaction between the two Asp residues prevents dimerization. In the absence of the negatively charged residue at position 154 (Asp → Ala) dimerization is improved. Fnr-I indicates an Ile to Ala substitution whereas Fnr-E indicates a Glu to Ala change in the H. seropedicae Fnr proteins. (C) β-Galactosidase activity from the BACTH assays using Fnr1 and Fnr3 proteins carrying point mutations at positions relative to 151 and 154 (respective to the E. coli protein).The graph is shaded in different colours to facilitate identification of the different sets of interactions tested. β-galactosidase assays were performed using cultures grown under oxygen-limiting conditions. Error bars show the standard deviation of three independent biological replicates carried out for each pairwise combination.