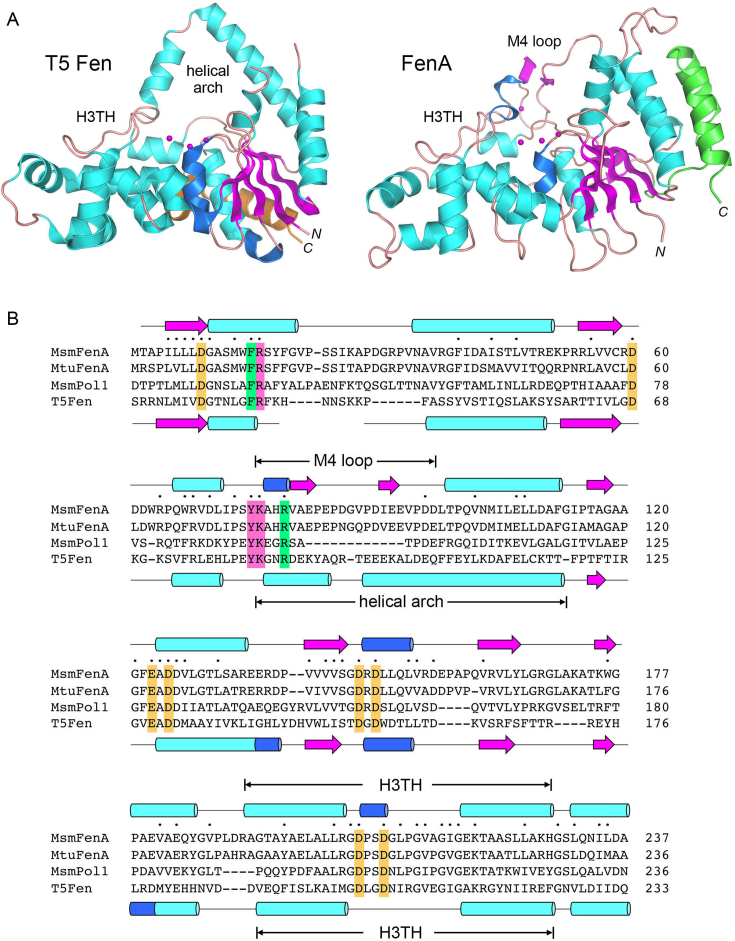
Figure 2.
Comparison of mycobacterial FenA to T5 Fen. (A) Side-by-side superposition of the structures of bacteriophage T5 Fen with three Mg2+ ions in the active site (at left, from accession code: 5HMM) and FenA with three Mn2+ ions in the active site (at right). Cartoon models of the tertiary structures are shown colored according to secondary structure, except for the two C-terminal α helices that are colored gold in T5 Fen and green in FenA to highlight their different spatial locations. The N and C termini are labeled in italics. The conserved DNA-binding H3TH motifs are labeled. The helical arch of T5 Fen is not conserved in FenA, which has a distinctive M4 loop as shown. (B) The amino acid sequence of Mycobacterium smegmatis (Msm) FenA from aa 1 to 237 is aligned to the corresponding sequences of the homologous Mycobacterium tuberculosis FenA (aa 1 to 236), the N-terminal 5′ exonuclease domain of M. smegmatis Pol1 (aa 18–233) and bacteriophage T5 Fen (aa 3–233). Gaps in the alignment are denoted by dashes. Positions of side chain identity/similarity in all four polypeptides are denoted by dots above the MsmFenA sequence. Secondary structure elements of FenA and T5 Fen (colored as in Figure 1) are shown above and below their amino acid sequences. Amino acids subjected to mutational analysis in the present study are highlighted in shaded boxes: gold for amino acids coordinating metals M1, M2 and M3; magenta for nearby amino acids in the active site; green for amino acids that, in T5 Fen, make contacts to DNA.