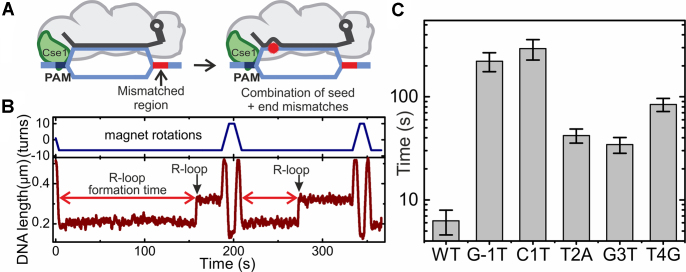
Figure 4.
Cascade binding kinetics measured on the shortened targets. (A) Sketch of Cascade bound to the DNA targets containing 6 PAM distal mismatches that prevent locking of the shortened 26-bp long R-loop. (B) Representative trajectory from magnetic tweezers measurement of repetitive R-loop formation-dissociation events obtained with E. coli Cascade on the shortened target. Jumps corresponding to R-loop formation are indicated. Dissociation of R-loops is obtained as soon as positive supercoiling (positive turns) is applied. The R-loop formation time is measured from the moment the desired negative supercoiling was reached. (C) Average R-loop formation times for the different protospacer/PAM variants obtained from exponential fits to of the recorded kinetics (Supplementary Figure S1). R-loop formation for seed and PAM mutants is at least five times slower than for the WT target.