Abstract
The expression of microRNA (miR-433) is altered in various types of human cancer. The present study analyzed the prognostic and biological value of miR-433 expression in colorectal cancer using reverse transcription-quantitative polymerase chain reaction in 125 colorectal tissue specimens (including a test cohort of 40 cases of paired colorectal cancer and adjacent normal mucosae and a confirmation cohort of 85 cases of stage I–III colorectal cancer). In vitro and nude mouse xenograft experiments were subsequently used to assess the effects of miR-433 expression on the regulation of colorectal cancer cell proliferation, adhesion, migration, and invasion. The data indicated that miR-433 expression was significantly downregulated in colorectal cancer tissues in the test and confirmation patient cohorts and that low miR-433 expression was associated with advanced tumor stage and early relapse. Furthermore, the restoration of miR-433 expression was able to significantly inhibit the proliferation of tumor cells by inducing G1-S cell cycle arrest, suppressing cyclinD1 and CDK4 expression, and markedly inhibited the migratory and invasive capacities of tumor cells in vitro. The restoration of miR-433 expression or liposome-based delivery of miR-433 mimics suppressed the growth of colorectal cancer cell xenografts in nude mice. In conclusion, miR-433 may be a putative tumor suppressor in colorectal cancer, and the detection of low miR-433 expression will be investigated in further studies as a putative biomarker for the detection of early relapse in patients with colorectal cancer.
Keywords: miR-433, tumor suppressor, colorectal cancer, early relapse
Introduction
Colorectal cancer is a major worldwide health burden due to high rates of cancer-associated morbidity and mortality. The most recent available data indicates that colorectal cancer accounts for approximately 1.4 million new cases of cancer and 693,900 mortalities globally in 2012 (1). Advancements in early detection using cancer screening tests and endoscopy have improved the prognosis of colorectal cancer and the development of novel treatment options (including minimally invasive surgical resection) allows effective control of tumor lesions. However, up to 38% of patients with early colorectal cancer following surgical resection develop tumor recurrence (2). Therefore, effective control of tumor recurrence (relapse) can reduce the rate of mortality due to colorectal cancer and improve the quality of life.
An estimated 40–50% of cases of tumor recurrence occur in the first year following initial surgery (3). The development of novel biomarkers may assist physicians to monitor and detect colorectal cancer relapse, while a novel class of targeting agents may help to further reduce the rate of tumor relapse. For example, the level of carcinoembryonic antigen (CEA) is frequently used as a clinical biomarker to survey colorectal cancer recurrence (4–6). However, there is no established biomarker or indicator to predict early relapse (7–9). Moreover, different targeting agents, including bevacizumab and cetuximab, which are inhibitors of vascular endothelial growth factor (VEGF) and epidermal growth factor receptor (EGFR), respectively, have been used in the clinic to treat colorectal cancer (10,11). However, the long-term survival rate remains very low for metastatic colorectal cancer (12). Taken together, there is an urgent need to improve the current understanding on the molecular mechanisms that underlie the development, progression and recurrence of colorectal cancer in order to develop novel strategies to improve the treatment of colorectal cancer in the clinic.
On a molecular level, it is widely accepted that the pathogenesis of colorectal cancer involves genetic alterations, including APC, K-RAS, and p53 (13). More recently, alteration of microRNA (miRNA) expression in human cancer, including colorectal cancer, has been thought to contribute to human carcinogenesis (14,15). These short and non-coding miRNA molecules are able to post-transcriptionally regulate the expression of their target genes (16) and can in turn regulate a variety of cell processes and homeostasis, including proliferation, apoptosis and differentiation in normal cells and invasion, migration and epithelial-mesenchymal transition (EMT) of tumor cells (17,18). For example, altered expression of miR-433 has been reported in numerous diseases. miR-433 functions as a potent inhibitor of migration of hepatocellular carcinoma (HCC) cells by directly targeting the cAMP response element binding protein (19). The expression of miR-433 was also downregulated in the visceral adipose tissues of patients with non-alcoholic steatohepatitis (20), hepatitis B virus-associated HCC (21) and human gastric cancer (22). Nevertheless, the ectopic expression of miR-433 was reported to inhibit the proliferation of gastric cancer cells by directly targeting the mRNA that encodes the oncogene, K-RAS (23). In non-neoplastic diseases, the expression of miR-433 inhibited BMP2-induced osteoblast differentiation by targeting Runx2 (24), while miR-433 was aberrantly expressed in myeloproliferative neoplasms and negatively regulates the proliferation and differentiation of CD34+ cells (25). Another study demonstrated that miR-433 also participated in TGF-β/Smad3-induced renal fibrosis and may be a potential therapeutic target in tissue fibrosis (26). Here, a pilot study was conducted to compare miR-433 expression in colorectal cancer tissues and paired normal mucosa tissues in order to investigate the association of miR-433 expression with clinicopathological data and tumor relapse.
The effects of miR-433 on the regulation of the malignant behaviors of colorectal cancer cells in vitro and in nude mouse xenografts were further investigated to assess whether miR-433 is a useful target for the control of colorectal cancer.
Materials and methods
Patients and tissue samples
In the present study, tissue samples were collected from two cohorts of patients with colorectal cancer for detection of miR-433 expression from January 2013 to May 2015. The age of the patients ranged from 42–76 years (mean, 62.3 years). The first cohort of patients included 40 patients with colorectal cancer, from whom normal and cancerous tissue samples were collected. The second cohort included 85 patients with stage I–III [The Union for International Cancer Control (UICC) staging system (27)] colorectal cancer. The latter cohort of patients included 40 early relapse patients and 45 patients without early relapse following radical tumor resection. The study was approved by the Institutional Review Board and Ethics Committee of the Nanfang Hospital of Southern Medical University, (Guangzhou, China). All patients that participated in the present study signed the informed consent form. Early relapse was defined as local recurrence (tumor growth within the primary site or restricted to the anastomosis) or distant metastasis (distant metastasis or diffuse peritoneal seeding) within 1 year following radical tumor resection according to previous studies (28,29). Patients with hereditary nonpolyposis colorectal cancer, familial adenomatous polyposis, or multiple primary cancers were excluded, while patients who received neoadjuvant treatment with either radiotherapy or chemotherapy prior to surgery were also excluded. All tissue samples were obtained from the operating room, immediately snap-frozen in liquid nitrogen and stored at −80°C until use. The present study was approved by The Ethics Committee of Nanfang Hospital (Guangzhou, China), and each patient provided written informed consent prior to participating in the present study or future research.
Cell lines and culture
Human colorectal cancer cell lines, SW480, HCT116, SW620 and LoVo, and a human normal colorectal mucosal cell line (FHC) were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and cultured in RPMI-1640 (Hyclone, Logan, UT, USA) or F12K (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) medium supplemented with 10% fetal bovine serum (FSB; Invitrogen; Thermo Fisher Scientific, Inc.) in a humidified incubator with 5% CO2 at 37°C.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total cellular RNA was isolated from tissue samples and cell lines FHC, SW480, HCT116, SW620 and LoVo using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Following quantification, the RNA samples were reverse transcribed (37°C for 15 min, 85°C for 5 sec) into cDNA using a PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd., Dalian, China), and the resulting cDNA was subjected RT-qPCR for detection of miR-433 expression using SYBR-Green PCR Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in an Applied Biosystems 7500 fast instrument according to the manufacturer's protocol. The sequences of the primers for miR-433 are as follows: Forward, 5′-AAGGCGCCTGAGGGAGGCACCACATCATCAGAT-3′ and reverse, 5′-TAAAGATCTGGCAGCCATCCTCGTGCTACTG-3′. U6 primers were used as a loading control and the sequences are as follows: Forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse, 5′-CGCTTCACGAATTTGCGTGTCAT-3′. The PCR thermocycling conditions were as follows: 95°C for 30 sec, followed by 40 cycles of 95°C for 5 sec, 60°C for 34 sec and 95°C for 15 sec; 60°C for 60 sec and 95°C for 15 sec. The experiment was repeated three times. The relative quantification of miR-433 level was calculated using the 2−ΔΔCq method (30), and the fold change of miR-433 was calculated using the 2−ΔΔCq equation. High miR-433 expression was defined as ≥0.498.
Transient and stable miRNA transfection
To restore miR-433 expression in colorectal cancer cells, hsa-miR-433 mimics were obtained from Applied Biosystems (Thermo Fisher Scientific, Inc.) and then transiently transfected into colorectal cancer cells using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) at a final concentration of 50 nM. To verify the effects of miR-433 and transfection efficiency, the pre-miR miRNA Precursor Molecules Negative Control from Applied Biosystems (Thermo Fisher Scientific, Inc.) was utilized as a control. A miR-433 expression vector was constructed for stable miR-433 expression in colorectal cancer lines by amplification, and a DNA fragment containing the full-length coding region of miR-433 cDNA (accession no. MI0001723) was cloned into the pcDNA3.1 vector (Addgene, Inc., Cambridge, MA, USA). After confirmation of the DNA sequence by sequencing, 1 µg plasmids were stably transfected into colorectal cancer cell lines using Lipofectamine® 2000 (Invitrogen), and miR-433 expression was assessed using RT-qPCR 48 h following transfection.
Cell proliferation CCK-8 and 5-ethynyl-2′-deoxyuridine labeling (EdU) assays
The proliferation ability of LoVo and SW620 tumor cells was assessed using a Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Shanghai, China). Briefly, tumor cells following miR-433 transfection were seeded into 96-well plates at a density of 5×103/well and cultured for 7 days at 37°C. Every 24 h, the cells were treated with CCK-8 at 37°C, according to the manufacturer's protocol and analyzed using a FACScan cytofluorimeter (BD Biosciences, Franklin Lakes, NJ, USA). To assess DNA synthesis, colorectal cancer cells were subjected to the EdU labeling assay. Specifically, following miR-433 transfection, tumor cells were seeded into 96-well plates at a density of 5×103/well and cultured for 7 days at 37°C. Every 24 h, the cells were subjected to EdU labeling and analyzed using a FACScan cytofluorimeter (Becton Dickinson). The experiment was repeated three times.
Tumor cell adhesion and cell cycle assays
In order to analyze the cell adhesion ability of tumor cells, tumor cells were seeded into 96-well plates that were pre-coated with 10 µg/ml fibronectin at 4°C for 18 h and allowed to adhere at 37°C for 90 min. At the end of this time period, adherent cells were quantified using the CCK-8 kit. To detect cell cycle distribution following miR-433 transfection, the cells were grown and transiently transfected with miR-433 mimics for 48 h and subsequently stained with propidium iodide (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and analyzed using a FACScan cytofluorimeter (Becton Dickinson) and the Cell Quest software (version 3.3; BD Biosciences) according to the manufacturer's instructions.
Transwell migration and invasion assays
Transwell inserts (pore size, 8 mm) were obtained from BD Biosciences and placed into 24-well cell culture plates. Colorectal cancer cells at a density of 2×105/ml in cell growth medium were added to the upper chamber in serum-free RPMI-1640 and the bottom chambers were filled with RPMI-1640 containing 20% FBS. After incubation at 37°C for 24 h, the non-migrated cells that remained on the upper surface of the membrane were scraped with a cotton swab, whereas the migrated cells on the lower face of the membrane were fixed with ice-cold methanol, stained with 5% Giemsa stain solution (Sigma-Aldrich; Merck KGaA), and counted under a light microscope from 10 random fields at a magnification of 200X for each triplicate sample. The tumor cell invasion assay was performed in a Transwell insert that was pre-coated with Matrigel, using the same conditions as the migration assay.
Tumor cell wound-healing assay
The cells were seeded at a density of 1×106 and transiently transfected with miR-433 mimics for 48 h at 37°C and then subjected to the wound-healing assay. In brief, after tumor cells had formed a monolayer, a wound across the dish was created by scratching with a 10 µl micropipette tip and then the cells were washed briefly with ice-cold phosphate-buffered saline (PBS) and further cultured for up to 48 h at 37°C. At the end of the experiment, images of the cells were captured using an Olympus IX53 microscope (Olympus Corporation, Tokyo, Japan) to assess wound closure.
Nude mouse colorectal cancer cell xenograft assay
The animal experiments were performed in compliance with the guidelines of the Ethics Committee of Medical Research, Southern Medical University. A total of 12 male BALB/c nude mice (weight, 18–22 g; age, 5–6 weeks) were housed in specific pathogen free laboratory conditions at 20–26°C and 55±5% humidity, 12 h light/dark cycle, and ad libitum access to food and water. To establish a mouse tumor cell xenograft model, 1 µg miR-433 plasmid or negative control plasmid was stably transfected into LoVo cells using Lipofectamine® 2000. LoVo cells (2×105 cells/mouse) were then injected subcutaneously into nude mice. After 40 days, the mice were sacrificed, and all tumor xenografts were collected for pathological exanimation. For miR-433 treatment, mice were treated by intratumoral injection of liposome-based miR-433 mimics and negative control at 10, 15, 20, 25 and 30 days. miR-433 mimics and negative control were injected at 4 spots around the tumor at a total volume of 50 µl and the final injection concentration used each time was 100 nM. miR-433 mimics (miR-433) and negative control (NC) were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). miR-433 sequence was AUCAUGAUGGGCUCCUCGGUGU and NC sequence was UUCUCCGAACGUGUCACGUTT. Tumor volume was estimated with the following formula: Length × width2 x0.5.
Protein extraction and western blot analysis
The cells were lysed in radioimmunoprecipitation buffer (Sigma-Aldrich; Merck KGaA), and protein concentration was quantified using a bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology, Guangzhou, China). These protein samples were resolved using 8–10% SDS-PAGE and transferred onto polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA). For western blotting, the membranes were incubated in 5% skimmed milk/PBS solution for 1 h at room temperature and then rinsed with Tris-buffered saline with 0.1% Tween (TBST) thrice prior to incubation with primary antibodies against PCNA (1:200; catalog no. ab29), fibronectin (1:4,000; catalog no. ab32419) (both from Abcam, Cambridge, MA, USA), cyclin D1 (1:1,000; catalog no. 2922), CDK4 (1:1,000; catalog no. 12790) (both from Cell Signaling Technology, Inc., Danvers, MA, USA), p21 (1:500; catalog no. 271532; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), E-cadherin (E-ca; 1:1,000; catalog no. 3195), N-cadherin (N-ca; 1:1,000; catalog no. 13116), vimentin (VIM; 1:1,000; catalog no. 5741) (all from Cell Signaling Technology, Inc.) and GAPDH (1:500; Santa Cruz Biotechnology Inc.) at 4°C overnight. The membranes were washed three times with TBST. The membranes were subsequently incubated with biotinylated anti-mouse IgG secondary antibody (1:20,000; catalog no. 14709; Cell Signaling Technology Inc.) and subsequently with an enhanced chemiluminescence solution (Pierce; Thermo Fisher Scientific, Inc.). Protein expression levels were quantified from band density using ImageJ software (version 1.38; National Institutes of Health, Bethesda, MD, USA) against the levels of GAPDH (loading control). The experiment was repeated three times.
Statistical analysis
The data are expressed as the mean ± standard deviation unless indicated otherwise. The unpaired Student's t-test was performed to compare miR-433 expression between test and control samples. Pearson's χ2 test was used to analyze the association of miR-433 expression with clinicopathological features from patients. The Kolmogorov-Smirnov test was used to test for normality in data from different groups. One-way analysis of variance was used to compare between different groups followed by Fisher's Least Significant Difference test for pairwise comparisons between groups. Two-way ANOVA was used to compare between groups in the xenograft experiments, followed by LSD test for pairwise comparisons between groups. All statistical analyses were conducted using GraphPad Software, (version 5.0; GraphPad Software, Inc., La Jolla, CA, USA) and SPSS software (version 18.0; SPSS Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Downregulation of miR-433 expression in colorectal cancer tissues
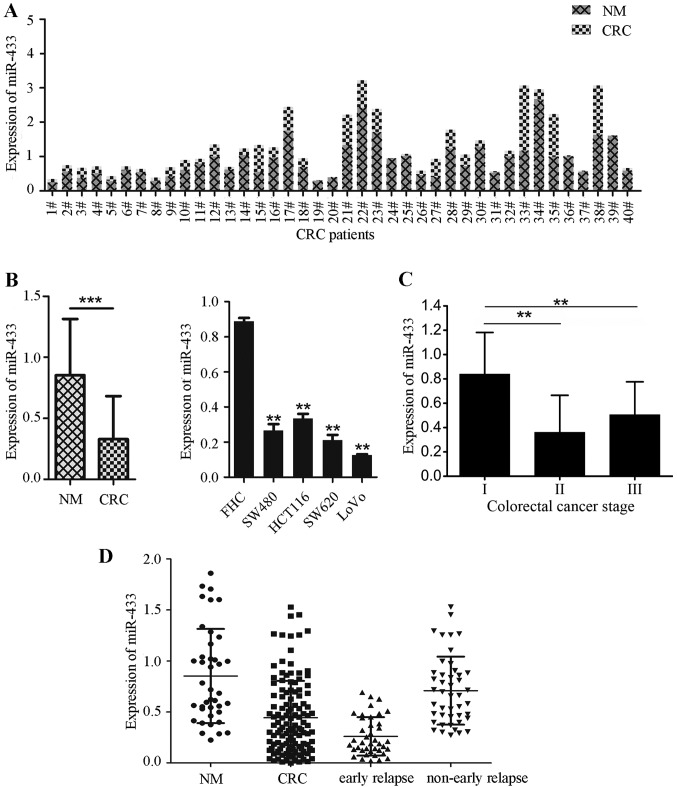
In the present study, miR-433 expression was analyzed in 40 pairs of frozen colorectal cancer and adjacent normal mucosa samples, and in four colorectal cancer cell lines (SW480, HCT116, SW620, and LoVo) and a human normal colorectal mucosal cell line (FHC) using RT-qPCR. The data indicated that miR-433 expression was significantly downregulated in colorectal cancer tissues compared with adjacent normal mucosae. Specifically, miR-433 expression was lower in 92.5% (37/40) of colorectal cancer tissues compared with the adjacent normal mucosae (Fig. 1A). By contrast, miR-433 expression was significantly decreased in colorectal cancer cell lines compared with FHC cells (all P<0.001; Fig. 1B). Out of the cell lines investigated, the lowest levels of miR-433 expression were observed in metastatic colorectal cancer SW620 and LoVo cell lines (Fig. 1B).
Figure 1.
Reduced miR-433 expression in human colorectal cancer tissue samples and cell lines. (A) RT-qPCR analysis of miR-433 levels. A total of 40 pairs of colorectal cancer and normal mucosae (NM) tissues were subjected to RT-qPCR analysis. ***P<0.0001. (B) RT-qPCR. Colorectal cancer cell lines and a normal cell line (FHC) were grown and then subjected to RT-qPCR analysis of miR-433 levels. **P<0.001 vs. FHC cells (one-way ANOVA and Fisher's Least Significant Difference test). (C) Mean miR-433 expression levels in colorectal cancer tissues at different stages of disease. **P<0.005 vs. stage I. Stage I, n=15; Stage II, n=31; Stage III, n=39; One-way ANOVA and Fisher's Least Significant Difference test. (D) Side-by-side comparison of miR-433 expression levels between colorectal cancer and normal mucosae tissues and between subjects with and without early-relapse. ANOVA, analysis of variance; CRC, colorectal cancer; miR, microRNA; NM, normal mucosae tissues; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Downregulated miR-433 expression is associated with early relapse of colorectal cancer
It was subsequently investigated whether miR-433 expression was associated with clinicopathological data and outcomes in the confirmation set of 85 colorectal cancer patients. The data indicated that the expression of miR-433 was significantly associated with tumor UICC stage when advanced UICC stages were compared with early UICC stages (P<0.01; Fig. 1C; Table I) and early relapse (P<0.001; Fig. 1D). However, miR-433 expression was not associated with sex, age, tumor differentiation, tumor size and lymph node metastasis (Table I).
Table I.
Association of miR-433 expression with clinicopathological data and outcome of colorectal cancer patients (n=85).
| miR-433 expression | ||||
|---|---|---|---|---|
| Parameters | n | High | Low | P-valuea |
| Sex, n | 0.730 | |||
| Male | 47 | 19 | 28 | |
| Female | 38 | 14 | 24 | |
| Age (years) | 0.130 | |||
| <65 | 37 | 11 | 26 | |
| ≥65 | 48 | 22 | 26 | |
| UICC stage, n | <0.001 | |||
| I | 15 | 12 | 3 | |
| II | 31 | 12 | 19 | |
| III | 39 | 9 | 30 | |
| Lymph node metastasis, n | 0.700 | |||
| + | 39 | 16 | 23 | |
| − | 46 | 17 | 29 | |
| Tumor differentiation, n | 0.730 | |||
| Well | 20 | 9 | 11 | |
| Medium | 41 | 16 | 25 | |
| Poor | 24 | 8 | 16 | |
| Tumor size, n (cm) | 0.220 | |||
| <5 | 45 | 20 | 25 | |
| ≥5 | 40 | 13 | 27 | |
| Early relapse, n | <0.001 | |||
| Yes | 40 | 4 | 36 | |
| No | 45 | 29 | 16 | |
χ2 test. miRNA, microRNA; UICC, Union for International Cancer Control.
Expression of miR-433 mimic inhibits the proliferation and adhesion ability of tumor cells to fibronectin, and promotes cell cycle arrest
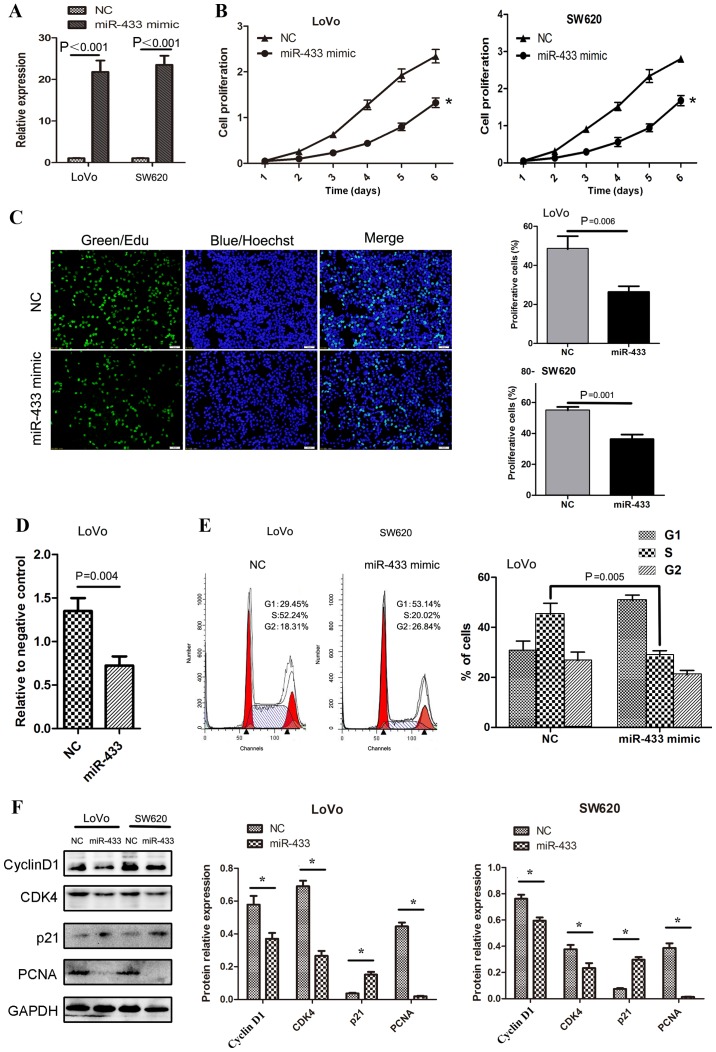
In order to gain an improved understanding of the role of miR-433 in regulating the development and progression of colorectal cancer, the effects of restoring miR-433 expression on the regulation of malignant behavior were assessed in colorectal cancer cells. Specifically, miR-433 mimics were transiently transfected into colorectal cancer cell lines, SW620 and LoVo. As indicated in Fig. 2A, the transfection of miR-433 mimics successfully restored miR-433 expression in these two cell lines, and the proliferation rate of these miR-433 transfected cell lines was significantly reduced (Fig. 2B and C). Furthermore, the adhesion ability of tumor cells to fibronectin was also inhibited following the overexpression of miR-433 in LoVo cells (P=0.004; Fig. 2D).
Figure 2.
Effects of miR-433 expression on the regulation of proliferation of colorectal cancer cells. (A) CCK-8 assay for assessing cell proliferation. Colorectal cancer cells were grown and transiently transfected with miR-433 mimics or negative control and then subjected to CCK-8 assay. *P<0.001 vs. NC; Two-way analysis of variance and Fisher's Least Significant Difference test. (B) Reverse transcription-quantitative polymerase chain reaction analysis confirmed successful restoration of miR-433 expression with miR-433 mimics. (C) EdU cell-labeling assay. LoVo cells were grown and transiently transfected with miR-433 mimics or negative control and then subjected to EdU labeling assay. Scale bar, 50 µm. (D) Tumor cell adhesion assay. Colorectal cancer LoVo cells were grown and transiently transfected with miR-433 mimics or negative control and then subjected to tumor cell adhesion assay. (E) Flow cytometric analysis of cell cycle. Colorectal cancer cells LoVo were grown and transiently transfected with miR-433 mimics or negative control and then subjected to flow cytometric assay for assessment of cell cycle. Graph indicating cell cycle arrest of at G1-S transition induced by miR-433 (P=0.005). (F) Altered expression of proteins that are associated with cell cycle as analyzed by western blotting. CDK4, cell dependent kinase 4; EdU, 5-ethynyl-2′-deoxyuridine; miR, microRNA; NC, negative control; NM, normal mucosae tissues; PCNA, proliferating cell nuclear antigen. *P<0.05 with comparisons shown by lines.
Furthermore, the effect of miR-433 expression on cell cycle distribution was investigated using a flow cytometer, and it was indicated that the percentage of cells in the G1 phase was markedly increased following the expression of miR-433 mimic, and that the percentage of cells in the S phase were significantly decreased following the expression of miR-433 mimic (Fig. 2E). Western blot analysis indicted that the cells that were arrested at G1 following the expression of miR-433 mimic exhibited significantly reduced expression of cyclin D1 and CDK4 proteins, which are key regulators of the G1 phase (P<0.05; Fig. 2F). However, the expression of the G1 phase inhibitor p21 was significantly increased following the expression of miR-433 mimic (P<0.05; Fig. 2F). The decrease in the proportion of cells in the S phase was confirmed by the significantly reduced expression of proliferating cell nuclear antigen (PCNA), which is a proliferation marker in cells that overexpress miR-433 (P<0.05; Fig. 2F).
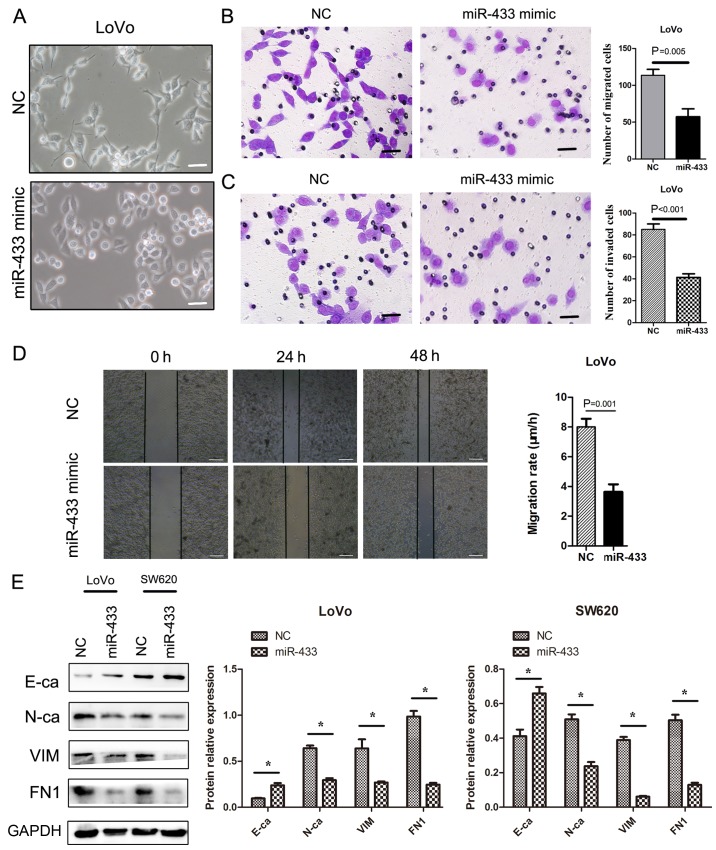
Expression of miR-433 mimic inhibits the migration and invasion of tumor cells in vitro
The restoration of miR-433 expression in LoVo cells resulted in cells exhibiting a rounded, cobblestone, epithelial-like morphology and more elongated cytoplasmic protuberances (Fig. 3A). Moreover, the expression of miR-433 mimic also significantly inhibited the migratory (P=0.005; Fig. 3B) and invasive capacities of tumor cells compared with control cells (P<0.001; Fig. 3C). In addition, the restoration of miR-433 expression also significantly suppressed the migration rate of the tumor cells in the wound-healing assay (P=0.001; Fig. 3D).
Figure 3.
Effects of miR-433 expression on the migration and invasion of colorectal cancer cells. (A) Morphology of tumor cells. Colorectal cancer LoVo cells were grown and transiently transfected with miR-433 mimics or negative control. The cells were subsequently viewed under a bright-phase microscope. The data indicated following transfection of miR-433 mimic, LoVo cells exhibited a rounded/cobble-stone morphology. By contrast, LoVo cells exhibited a more elongated morphology. Scale bar, 50 µm. (B) Transwell migration assay. Colorectal cancer LoVo cells were grown and transiently transfected with miR-433 mimics or negative control. The cells were subjected to Transwell assay. Scale bar, 50 µm. (C) Transwell invasion assay. Colorectal cancer LoVo cells were grown and transiently transfected with miR-433 mimics or negative control and then subjected to the Transwell invasion assay. Scale bar, 50 µm. (D) Wound-healing assay. Colorectal cancer LoVo cells were grown and transiently transfected with miR-433 mimics or negative control and then subjected to a wound-healing assay. Scale bar, 200 µm. (E) Western blot results. Colorectal cancer LoVo cells were grown and transiently transfected with miR-433 mimics or negative control and then subjected to western blot analysis for E-cadherin, N-cadherin, vimentin, and fibronectin. E-ca, E-cadherin; FN1, fibronectin; N-ca, N-cadherin; VIM, vimentin; miR, microRNA; NC, negative control. *P<0.05 with comparisons shown by lines.
Furthermore, western blot analysis indicated that ectopic miR-433-expressing colorectal cancer cells exhibited significantly upregulated E-cadherin expression but significantly downregulated N-cadherin, VIM and fibronectin (FN1) expression compared with negative control (P<0.05; Fig. 3E).
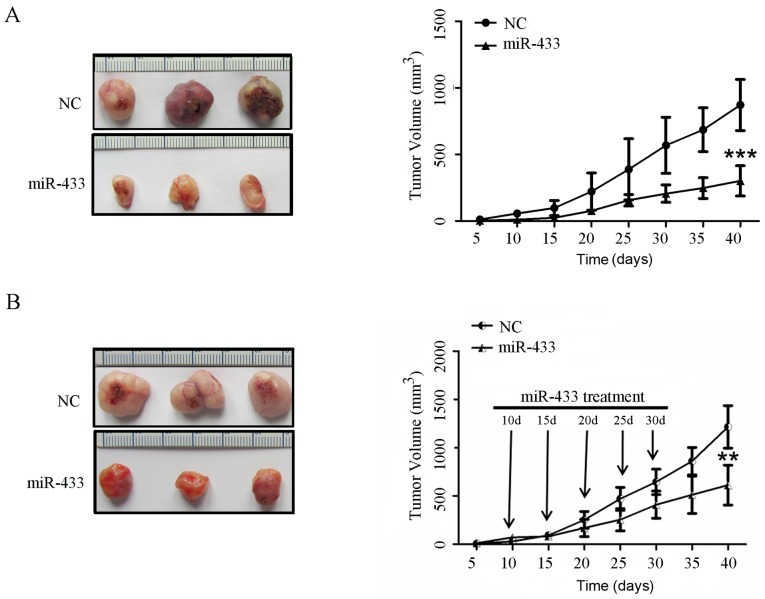
Effects of restored miR-433 expression on suppression of colorectal cancer cell xenograft growth in nude mice
In order to validate the role of miR-433 in vivo, an in vivo nude mouse xenograft assay was performed. A stable miR-433-expressing LoVo cell subline was first established, and the cells were injected into nude mice. The data indicated that the mice that were implanted with miR-433-overexpressing colorectal cancer cells exhibited significantly slower tumor growth after 40 days compared with the animals that were injected with negative control cells (P<0.001; Fig. 4A).
Figure 4.
Effects of miR-433 on the growth of tumor cell xenografts in nude mice. (A) Colorectal cancer cell nude mouse xenograft model. The restoration of miR-433 expression inhibited subcutaneous tumor xenograft growth in nude mice. The tumor masses were smaller in mice that were injected with miR-433-expressing cells compared with mice that were injected with negative control cells. (B) miR-433 therapy in nude mice with colorectal cancer cell xenografts. The growth of tumor cell xenografts was suppressed following intratumoral injection of liposomes containing miR-433 mimics compared with NC at 10, 15, 20, 25 and 30 days following LoVo cell implantation. Tumor volumes are presented as the mean ± standard deviation. Growth curves were compared using two-way analysis of variance and Fisher's Least Significant Difference test. **P<0.001, ***P<0.0001 vs. NC. NC, negative control.
Furthermore, LoVo cell xenografts in nude mice were established and then treatment was initiated by intratumoral injection of liposome-based miR-433 mimics and negative control at 10, 15, 20, 25 and 30 days. The data showed that the mice that were treated with miR-433 mimics exhibited a significant reduction in tumor xenograft growth compared with the control treatment (P<0.001; Fig. 4B).
Discussion
Colorectal cancer relapse following surgical resection is an important factor leading to poor survival in colorectal cancer patients (31). The identification and evaluation of molecular markers, such as miRNAs, may assist in the diagnosis of patients with potential early tumor relapse. These miRNAs are stable in tissues and cells and are not susceptible to degradation. These characteristics make miRNAs a better biomarker in the clinic because the altered expression of various miRNAs has been reported to predict cancer development, progression, and prognosis (32,33). miR-433 expression was reported to be absent or reduced in various types of human cancer (19,22,34). Therefore, in the present study, the expression of miR-433 was determined in colorectal cancer tissues and paired normal mucosae. It was then determined whether miR-433 expression was associated with clinicopathological data from patients with colorectal cancer using two different cohorts. Subsequently, the role of miR-433 was determined in vitro and in a nude mouse xenograft model. It was detected that miR-433 expression was lower in colorectal cancer tissues compared with adjacent normal tissues.
A low expression of miR-433 was associated with advanced tumor stages and early relapse in patients with colorectal cancer. Furthermore, the in vitro and nude mouse data indicated that restoring miR-433 expression in colorectal cancer cell lines was able to decrease cell proliferation, migration and invasion in vitro. Restoring miR-433 expression was also able to reduce the growth of colorectal cancer cell xenografts.
miRNAs function to regulate and regulate a large number of cellular processes in the human body (35) and have a high degree of sequence conservation among distantly related organisms, indicating that they participate in essential biological processes in different organisms (36). In the present study, the data indicated reduced miR-433 expression in colorectal cancer tissues compared with adjacent normal tissues, which was associated with advanced tumor stages and early relapse. These data suggest that miR-433 has an antitumor effect on colorectal cancer and may be a putative tumor suppressor gene in colorectal cancer. In the literature, it was reported that miR-433 expression was also reduced in gastric cancer (22,23), hepatocellular carcinoma (21) and oral squamous cell carcinoma (37). However, in ovarian cancer cells, the overexpression of miR-433 either induced the resistance of tumor cells to paclitaxel (34) or negatively regulates the expression of thymidylate synthase which mediates 5-fluorouracil sensitivity in HeLa cells (38). Therefore, these data indicated that miR-433 may function differently in different types of human cancer, and the further study of the miR-433-regulated pathways may help to elucidate the functions and role of miR-433 in human cancer.
Furthermore, in the present study the potential mechanisms of miR-433 in the inhibition of colorectal cancer relapse were investigated using in vitro experiments. It was detected that transfection with miR-433 mimic significantly reduced the proliferation of colorectal cancer cells, potentially by arresting the cells at G1-S transition. Furthermore, the migratory and invasive capacities of tumor cells were inhibited following transfection with miR-433 mimic, which is consistent with data reported for other types of cancer (37). At the genetic level, transfection with miR-433 mimic suppressed the levels of cyclin D1 and CDK4 proteins compared with negative control. Indeed, the cyclin D1/CDK4 complex is a key regulator of cell cycle transition through the G1 phase and governs the cell cycle progression (39). The restoration of miR-433 expression also increased p21 levels, which, in turn, inhibited cells from entering the S phase for duplication of chromosomes. The p21 protein has been reported to have an important role in the suppression of the G1/S cell cycle transition (40). Furthermore, the current data also demonstrated that the transfection of miR-433 mimic inhibited the migration and invasion of colorectal cancer cells, which further supported previous related studies on other cancer types (23,37). It was also observed that the transfection of miR-433 mimic increased E-cadherin expression but inhibited the expression of N-cadherin, VIM, and FN1 proteins, indicating that miR-433 may suppress EMT in colorectal cancer cells, and the latter has been previously reported as associated with cancer metastasis (41). The in vitro data in present study was then confirmed in the nude mouse xenograft model, using miR-433 expressing-colorectal cancer cells and treatment with miR-433 mimics.
In conclusion, the current study used ex vivo, in vitro and nude mouse xenograft models to demonstrate that the loss of miR-433 expression may contribute to the development and progression of colorectal cancer and that the restoration of miR-433 expression suppresses malignant behavior in tumor cells. However, further studies are required to investigate the clinical effects of miR-433 on patients with colorectal cancer. Moreover, miR-433 may be developed as a potential biomarker for the prediction of colorectal cancer relapse.
Acknowledgements
Not applicable.
Funding
The present study was supported in part by a grant from the Affiliated Hospital of Inner Mongolia Medical University (grant no. NYFY YB 2014005).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JZ, LZ and LH were involved in the conception and design of the study; JZ, LZ, TZ and XM were responsible for the acquisition of data; JZ, LZ, TZ, XM and YZ performed analysis and interpretation of data, for example statistical analysis, biostatistics, computational analysis; JZ and LZ were involved in the writing, review and revision of the manuscript; JZ and TZ provided administrative, technical and material support, for example reporting or organizing data and constructing databases. LH also supervised the study.
Ethics approval and consent to participate
The study was approved by the Institutional Review Board and Ethics Committee of The Nanfang Hospital of Southern Medical University, (Guangzhou, China). Written informed consent was obtained from all patients.
Consent for publication
All patients that participated in the present study signed the informed consent form.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Li X, Li X, Liao D, Wang X, Wu Z, Nie J, Bai M, Fu X, Mei Q, Han W. Elevated microRNA-23a expression enhances the chemoresistance of colorectal cancer cells with microsatellite instability to 5-fluorouracil by directly targeting ABCF1. Curr Protein Pept Sci. 2015;16:301–309. doi: 10.2174/138920371604150429153309. [DOI] [PubMed] [Google Scholar]
- 3.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 4.Duffy MJ. Carcinoembryonic antigen as a marker for colorectal cancer: Is it clinically useful? Clin Chem. 2001;47:624–630. [PubMed] [Google Scholar]
- 5.Duffy MJ, van Dalen A, Haglund C, Hansson L, Klapdor R, Lamerz R, Nilsson O, Sturgeon C, Topolcan O. Clinical utility of biochemical markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines. Eur J Cancer. 2003;39:718–727. doi: 10.1016/S0959-8049(02)00811-0. [DOI] [PubMed] [Google Scholar]
- 6.Hammarstrom S. The carcinoembryonic antigen (CEA) family: Structures, suggested functions and expression in normal and malignant tissues. Semin Cancer Biol. 1999;9:67–81. doi: 10.1006/scbi.1998.0119. [DOI] [PubMed] [Google Scholar]
- 7.Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 8.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 10.Macedo LT, da Costa Lima AB, Sasse AD. Addition of bevacizumab to first-line chemotherapy in advanced colorectal cancer: A systematic review and meta-analysis, with emphasis on chemotherapy subgroups. BMC Cancer. 2012;12:89. doi: 10.1186/1471-2407-12-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian S, Simon I, Moreno V, Roepman P, Tabernero J, Snel M, van't Veer L, Salazar R, Bernards R, Capella G. A combined oncogenic pathway signature of BRAF, KRAS and PI3KCA mutation improves colorectal cancer classification and cetuximab treatment prediction. Gut. 2013;62:540–549. doi: 10.1136/gutjnl-2012-302423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou SW, Huang YY, Wei Y, Jiang ZM, Zhang YD, Yang Q, Xie DR. No survival benefit from adding cetuximab or panitumumab to oxaliplatin-based chemotherapy in the first-line treatment of metastatic colorectal cancer in KRAS wild type patients: A meta-analysis. PLoS One. 2012;7:e50925. doi: 10.1371/journal.pone.0050925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 14.Szafranska AE, Davison TS, John J, Cannon T, Sipos B, Maghnouj A, Labourier E, Hahn SA. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 15.Hur K, Toiyama Y, Okugawa Y, Ide S, Imaoka H, Boland CR, Goel A. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut. 2017;66:654–665. doi: 10.1136/gutjnl-2014-308737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 17.Spizzo R, Nicoloso MS, Croce CM, Calin GA. SnapShot: MicroRNAs in cancer. Cell. 2009;137:586.e1. doi: 10.1016/j.cell.2009.04.040. [DOI] [PubMed] [Google Scholar]
- 18.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Z, Tsuchiya H, Zhang Y, Hartnett ME, Wang L. MicroRNA-433 inhibits liver cancer cell migration by repressing the protein expression and function of cAMP response element-binding protein. J Biol Chem. 2013;288:28893–28899. doi: 10.1074/jbc.M113.502682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Estep M, Armistead D, Hossain N, Elarainy H, Goodman Z, Baranova A, Chandhoke V, Younossi ZM. Differential expression of miRNAs in the visceral adipose tissue of patients with non-alcoholic fatty liver disease. Aliment Pharmacol Ther. 2010;32:487–497. doi: 10.1111/j.1365-2036.2010.04366.x. [DOI] [PubMed] [Google Scholar]
- 21.Wang W, Zhao LJ, Tan YX, Ren H, Qi ZT. Identification of deregulated miRNAs and their targets in hepatitis B virus-associated hepatocellular carcinoma. World J Gastroenterol. 2012;18:5442–5453. doi: 10.3748/wjg.v18.i38.5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo H, Zhang H, Zhang Z, Zhang X, Ning B, Guo J, Nie N, Liu B, Wu X. Down-regulated miR-9 and miR-433 in human gastric carcinoma. J Exp Clin Cancer Res. 2009;28:82. doi: 10.1186/1756-9966-28-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo LH, Li H, Wang F, Yu J, He JS. The tumor suppressor roles of miR-433 and miR-127 in gastric cancer. Int J Mol Sci. 2013;14:14171–14184. doi: 10.3390/ijms140714171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim EJ, Kang IH, Lee JW, Jang WG, Koh JT. MiR-433 mediates ERR gamma-suppressed osteoblast differentiation via direct targeting to Runx2 mRNA in C3H10T1/2 cells. Life Sci. 2013;92:562–568. doi: 10.1016/j.lfs.2013.01.015. [DOI] [PubMed] [Google Scholar]
- 25.Lin X, Rice KL, Buzzai M, Hexner E, Costa FF, Kilpivaara O, Mullally A, Soares MB, Ebert BL, Levine R, et al. miR-433 is aberrantly expressed in myeloproliferative neoplasms and suppresses hematopoietic cell growth and differentiation. Leukemia. 2013;27:344–352. doi: 10.1038/leu.2012.224. [DOI] [PubMed] [Google Scholar]
- 26.Li R, Chung AC, Dong Y, Yang W, Zhong X, Lan HY. The microRNA miR-433 promotes renal fibrosis by amplifying the TGF-β/Smad3-Azin1 pathway. Kidney Int. 2013;84:1129–1144. doi: 10.1038/ki.2013.272. [DOI] [PubMed] [Google Scholar]
- 27.Sobin LH, Gospodarowicz MK, Wittekind Ch. 7th. Wiley-Blackwell; Oxford, UK: 2009. International Union Against Cancer (UICC) TNM Classification of Malignant Tumors. [Google Scholar]
- 28.Tsai HL, Chu KS, Huang YH, Su YC, Wu JY, Kuo CH, Chen CW, Wang JY. Predictive factors of early relapse in UICC stage I–III colorectal cancer patients after curative resection. J Surg Oncol. 2009;100:736–743. doi: 10.1002/jso.21404. [DOI] [PubMed] [Google Scholar]
- 29.Kidner CA, Martienssen RA. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature. 2004;428:81–84. doi: 10.1038/nature02366. [DOI] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Li S, Gao J, Gu J, Yuan J, Hua D, Shen L. MicroRNA-215 inhibits relapse of colorectal cancer patients following radical surgery. Med Oncol. 2013;30:549. doi: 10.1007/s12032-013-0549-0. [DOI] [PubMed] [Google Scholar]
- 32.Nair VS, Maeda LS, Ioannidis JP. Clinical outcome prediction by microRNAs in human cancer: A systematic review. J Natl Cancer Inst. 2012;104:528–540. doi: 10.1093/jnci/djs027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hui A, How C, Ito E, Liu FF. Micro-RNAs as diagnostic or prognostic markers in human epithelial malignancies. BMC Cancer. 2011;11:500. doi: 10.1186/1471-2407-11-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiner-Gorzel K, Dempsey E, Milewska M, McGoldrick A, Toh V, Walsh A, Lindsay S, Gubbins L, Cannon A, Sharpe D, et al. Overexpression of the microRNA miR-433 promotes resistance to paclitaxel through the induction of cellular senescence in ovarian cancer cells. Cancer Med. 2015;4:745–758. doi: 10.1002/cam4.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 36.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 37.Wang XC, Ma Y, Meng PS, Han JL, Yu HY, Bi LJ. miR-433 inhibits oral squamous cell carcinoma (OSCC) cell growth and metastasis by targeting HDAC6. Oral Oncol. 2015;51:674–682. doi: 10.1016/j.oraloncology.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 38.Gotanda K, Hirota T, Matsumoto N, Ieiri I. MicroRNA-433 negatively regulates the expression of thymidylate synthase (TYMS) responsible for 5-fluorouracil sensitivity in HeLa cells. BMC Cancer. 2013;13:369. doi: 10.1186/1471-2407-13-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Datar SA, Galloni M, de la Cruz A, Marti M, Edgar BA, Frei C. Mammalian cyclin D1/Cdk4 complexes induce cell growth in Drosophila. Cell Cycle. 2006;5:647–652. doi: 10.4161/cc.5.6.2573. [DOI] [PubMed] [Google Scholar]
- 40.Harada K, Ogden GR. An overview of the cell cycle arrest protein, p21(WAF1) Oral Oncol. 2000;36:3–7. doi: 10.1016/S1368-8375(99)00049-4. [DOI] [PubMed] [Google Scholar]
- 41.Yates CC, Shepard CR, Stolz DB, Wells A. Co-culturing human prostate carcinoma cells with hepatocytes leads to increased expression of E-cadherin. Br J Cancer. 2007;96:1246–1252. doi: 10.1038/sj.bjc.6603700. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.