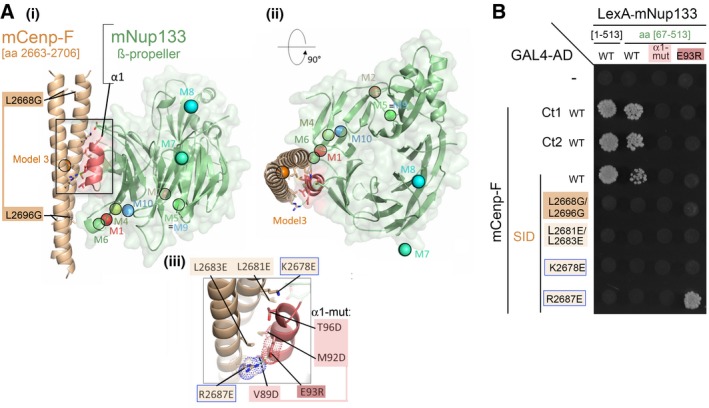
Predicted Model 3, involving interaction of mCenp‐F‐SID (parallel coiled‐coil, brown) with helix α1 (red) of mNup133 β‐propeller (green). Two orthogonal views are presented in panels (i) and (ii). In (i), Cenp‐F mutated residues impairing dimerization L2669G/L2696G are highlighted. (iii) presents a detailed view of the interface, highlighting the interaction between
mCenp‐FR2687 and
mNup133E93. Mutations designed to test this model are indicated as sticks and labeled in panels (i) and (iii). The colored spheres in (i) and (ii) represent the center of mass of Cenp‐F dimeric segments for the 10 most likely binding modes of Cenp‐F to Nup133 as predicted by InterEvDock server (M1 to M10)
41.