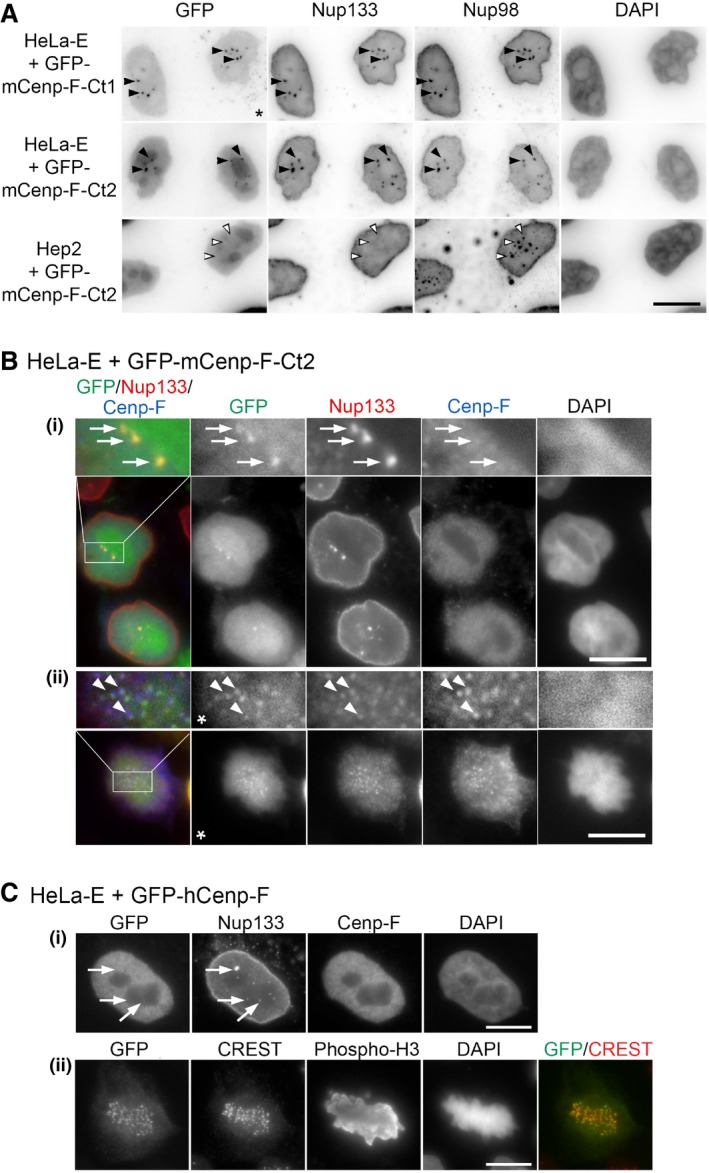
HeLa‐E cells transfected with GFP‐mCenp‐F‐Ct2 (B) or GFP‐hCenp‐F (C) were fixed 2 and 1 day after transfection, respectively, and immunolabeled with the indicated antibodies. A single plane is shown except for panels (C, ii) where a projection of 11 z‐sections is presented to better visualize the kinetochores (marked by the CREST serum). In (B), threefold magnifications of the marked areas are also presented. Arrows in the G2 cells shown in (B, (i)) and (C, (i)) (identified based on their strong Cenp‐F levels) point to GLFG bodies stained by anti‐Nup133 that contain GFP‐mCenp‐F‐Ct2 but neither endogenous Cenp‐F (the anti‐Cenp‐F antibody used here does not recognize the transfected GFP‐mCenp‐F‐Ct2 construct) nor GFP‐hCenp‐F. Arrowheads in the metaphase cell (B, ii) highlight co‐localization of GFP‐mCenp‐F‐Ct2, Nup133, and endogenous Cenp‐F at kinetochores. Note that a clear localization of the KT‐core domain of hCenp‐F was previously reported to require either the internal repeats or a longer C‐terminal extension including the C‐terminal CAAX farnesylation site that are absent from our construct
27,
28. This minor discrepancy likely reflects the improved detection due to our pre‐extraction conditions and the use of a GFP tag that does not rely on antibody detection. The star indicates that the gamma was altered on that image to improve the visualization of GFP‐mCenp‐F‐Ct2 kinetochore localization. Scale bars, 10 μm.