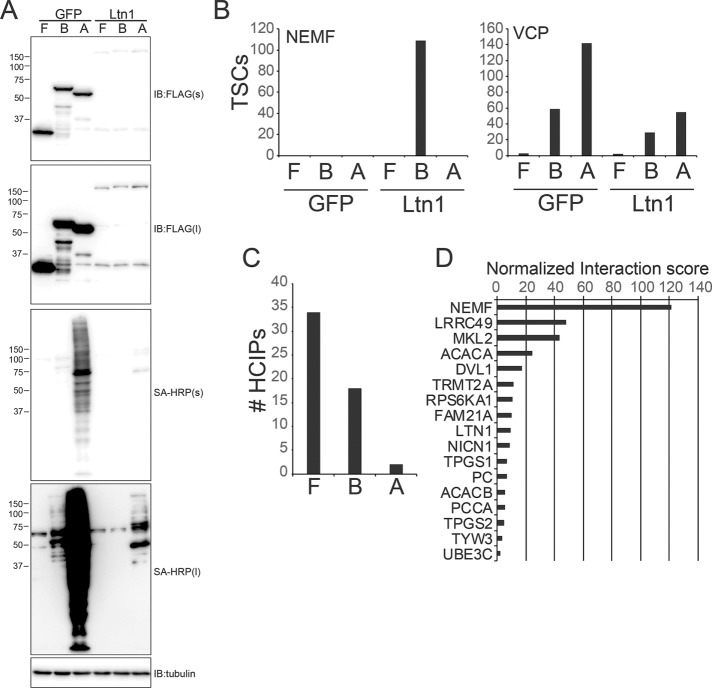
FIGURE 2:
Identification of Ltn1-interacting proteins using proximity labeling approaches. (A) 293 Flp-In cells with dox-induced expression of FLAG-HA(F)-, BirA*-FLAG(B)-, or FLAG-APEX2(A)–tagged GFP or Ltn1 were treated with doxycycline overnight, and protein biotinylation was induced as described for BirA*- or APEX2-tagged cell lines. Whole-cell extracts were immunoblotted with a-FLAG antibodies or probed with SA-HRP. Short (s) or long (l) exposures are depicted. (B) The summed total spectral counts (TSCs) from replicate experiments corresponding to NEMF (left) or VCP (right) from streptavidin or anti-FLAG–isolated and identified proteins from FLAG-HA(F)-, BirA*-FLAG(B)-, or FLAG-APEX2(A)–tagged GFP- or Ltn1-expressing cell extracts. For panels B–D, all IPs were performed in triplicate from distinct experiments and cell lysates. (C) The number of high-confidence interacting proteins (HCIPs) for Ltn1 for each of the three proteomic platforms was determined using a modified CompPASS scoring method. (D) The normalized interaction scores as determined by the CompPASS scoring method for top-scoring Ltn1 interacting proteins identified using the BioID approach is shown.