Abstract
Sucrose is a crucial compound for the growth and development of plants, and the regulation of multiple genes depends on the amount of soluble sugars present. Sucrose acts as a signaling molecule that regulates a proton-sucrose symporter, with its sensor being the sucrose transporter. Flavonoid and anthocyanin biosynthesis are regulated by sucrose, and sucrose signaling can affect flavonoid and anthocyanin accumulation. In the present study, we found a Myb transcription factor affecting accumulation of anthocyanin. AtMyb56 showed an increase in its expression in response to sucrose treatment. Under normal conditions, anthocyanin accumulation was similar between Col-0 (wild type) and atmyb56 mutant seedlings; however, under sucrose treatment, the level of anthocyanin accumulation was lower in the atmyb56 mutant plants than in Col-0 plants. Preliminary microarray analysis led to the investigation of the expression of one candidate gene, AtGPT2, in the atmyb56 mutant. The phosphate translocator, which is a plastidial phosphate antiporter family, catalyzes the import of glucose-6-phosphate (G-6-P) into the chloroplast. AtGPT2 gene expression was altered in atmyb56 seedlings in a sucrose-dependent manner in response to circadian cycle. Furthermore, the lack of AtMyb56 resulted in altered accumulation of maltose in a sucrose-dependent manner. Therefore, the sucrose responsive AtMyb56 regulates AtGPT2 gene expression in a sucrose-dependent manner to modulate maltose and anthocyanin accumulations in response to the circadian cycle.
Keywords: Anthocyanin, Arabidopsis thaliana, AtMyb56, Myb transcription factor, plastidial glucose-6-phosphate/phosphate translocator, sucrose
INTRODUCTION
Plants respond to various stress factors by modifying their metabolic pathways in different ways to survive the stress period and recover from the damage caused by the stress factors (Dos Reis et al., 2012). Plants have successfully adapted to environments via various mechanisms in response to a combination of different stress factors. To detect changes in the environment and respond to the complexity of stress conditions in a unique way, plants, which are sessile organisms, have evolved specific pathways to minimize damage and conserve precious resources for growth and reproduction (Rizhsky et al., 2004). In particular, metabolic and cellular reprogramming functions to control the pathways for regulation and signaling in response to stress (Wu and Jinn, 2012). Furthermore, plant secondary metabolites serve various health-related functions, for example, preventing cancer, cardiovascular disease, and diabetes owing to their role as free radical scavengers (Lee et al., 2016).
The MYB genes, which are found in all eukaryotes, constitute a large family of genes that encode proteins with diverse functions. MYB family genes are classified by the number of imperfect repeats within the MYB domain: R1R2R3R4 MYB, R1R2R3 MYB, R2R3 MYB, single repeat MYB, and the MYB-like type (Rosinski and Atchley, 1998; Jin and Martin, 1999; Dubos et al., 2010; Zhang et al., 2012). The majority of plant MYB genes belong to the R2R3 MYB type (Stracke et al., 2001). A DNA binding domain with a helix-turn-helix structure (Ogata et al., 1994) and C-terminal region that is highly variable and responsible for interacting with other components combine to form the MYB repeat (Dias et al., 2003; Matus et al., 2008). R2R3 MYB genes regulate many cellular processes including secondary metabolism and the determination of the fate of cells in plants (Stracke et al., 2001). In addition, MYB transcription factors play positive/negative roles in various signaling pathways. For instance, the cross-talk of abscisic acid (ABA)-auxin is mediated by AtMyb96, a molecular link, during the response to drought-stress and the growth of lateral roots (Seo et al., 2009). The changes in gibberellins (GA) metabolism and signaling are regulated by AtMyb62 in response to the depletion of phosphate (Devaiah et al., 2009). Overexpression of AtMyb15 possibly leads to the enhanced expressions of genes associated with ABA biosynthesis and improves the resistance of Arabidopsis under abiotic stresses (Ding et al., 2009). More than 125 R2R3 MYBs have been discovered in Arabidopsis; however, the functions of these transcription factors remain elusive (Yanhui et al., 2006).
Sugars play many important roles throughout the life cycle of plants (Smeekens, 2000; Rolland et al., 2002; Rook and Bevan, 2003), and are involved in a variety of signaling cascades, including nutrient mobilization that stimulates growth, photosynthesis, and flavonoid biosynthesis (Koch, 1996; Rolland et al., 2002). Among these roles, metabolic sugar sensing systems are conserved in plants (SNF1-related protein kinase 1; SnRK1), mammals (AMP-activated protein kinase; AMPK), and yeast (sucrose non-fermenting 1; SNF1), which possess highly conserved eukaryotic protein kinase families (Hardie, 2007; Polge and Thomas, 2007; Hedbacker and Carlson, 2008). However, detailed information regarding plant sugar signaling remains elusive, and the identification of many molecular components is required.
Anthocyanins, which are water-soluble flavonoids synthesized via the phenylpropanoid pathway, function as pigments for the formation of red, purple, and blue colors depending on pH concentration. Arabidopsis plants accumulate greater levels of anthocyanins in sucrose-treated medium (Tsukaya et al., 1991; Ohto et al., 2001). Furthermore, anthocyanins have various and crucial roles, including insect attractants and sun protectants in plants (Ross and Kasum, 2002). Moreover, they act as powerful antioxidants, thereby promoting health in humans (Khodabande et al., 2017; Yang et al., 2017). Therefore, the biosynthesis of anthocya-nin is dynamically controlled in response to a variety of environmental challenges, which includes transcriptional regulation of the corresponding genes. For example, anthocyanin biosynthesis is significantly enhanced in pears, apples, and red orange fruits when stored at low temperatures (Li et al., 2012; Lo Piero et al., 2005; Ubi et al., 2006). The biosynthesis of anthocyanin is regulated by diverse Myb transcription factors, including positive regulators (PAP1, PAP2, Myb113, and Myb114) and negative regulators (MybL2; Borevitz et al., 2000; Dubos et al., 2008; Gonzalez et al., 2008 ; Tohge et al., 2005). These findings indicate that Myb transcription factors serve crucial roles in controlling anthocyanin biosynthesis. In the present study, One Myb gene, AtMYB56, which is induced by sucrose and is a potent regulator of anthocyanin accumulation, was identified. We suggest AtMYB56 is a key regulator of sucrose-induced AtGPT2 expression in response to circadian cycle, and thereby alters the level of free maltose and the subsequent accumulation of anthocyanins in plants.
MATERIALS AND METHODS
Plant material and growth conditions
Plants (A. thaliana) were grown at 23°C under a day/night light regime of 16 h light/8 h dark and 60% relative humidity. The atmyb56 knockout mutants (atmyb56#1, SALK_060289; atmyb56#2, SALK_062413) were obtained from the Arabidopsis Biological Resource Center (Columbus, OH, USA) and A. thaliana Col-0 was utilized as the wild type (WT). To examine the growth and development of plants, seeds were plated onto Murashige and Skoog agar (2% sucrose, pH 5.7) media. For the extraction of RNA, plants were grown on 10 ml of 1 × Murashige and Skoog media supplemented with compounds specific to the treatment being investigated.
Anthocyanin measurements
Whole seedlings from 4-day-old plants were collected and pulverized in liquid nitrogen. Anthocyanins were extracted overnight with 80% methanol (v/v 5% HCl) at 4°C in the dark. The supernatant was collected after centrifugation at 800 × g for 2 min and placed in a new tube, and anthocyanin levels were quantified photometrically (PowerWave XS; Bio Tek Instruments, Inc.).
Construction of green fluorescent protein (GFP) fusion protein and AtMyb56-OE lines
The AtMyb56 coding region was amplified with reverse transcription polymerase chain reaction (RT-PCR) to generate AtMyb56 fused with GFP using total RNA extracted from A. thaliana (Col-0) seedlings and the following primers: AtMyb56-F, 5′-ATG AAT CCA AAT CTC CTT GAG AA-3′; and AtMyb56-R, 5′-GGA AGC TCC AAC TCC AAG AAA A-3′. The products of PCR were cloned into the pMDC83 gateway vector (Curtis and Grossniklaus, 2003). The particle bombardment method was applied to transfer ligated vector AtMyb56::GFP to onion epidermal cells. The same cDNA was cloned into a TOPO vector using a pCR™8/GW/TOPOR TA CloningR Kit (Invitrogen™, https://www.thermofisher.com/kr/ko/home/brands/invitrogen.html). Then, it was transferred into a destination pMDC32 vector (Curtis and Grossniklaus, 2003) to generate the 35S promoter:AtMyb56 overexpressing (OE) lines.
Flavonol staining
Four-day-old Col-0, atmyb56#1, atmyb56#2, and AtMyb56 OE line #2 seedlings grown on 2% sucrose media were transferred to a staining solution (saturated [0.25%, w/v] diphenylboric acid 2-aminoethyl ester [DPBA] with 0.005 % Triton X-100) and stained for 15 min. Confocal laser scanning microscopy was used to visualize flavonol localization and color. Fluorescence was measured using an LSM 5 Exciter (Carl-Zeiss Microscopy GmbH) with an EC Plan-NEOFLUAR 10×/0.30 objective lens, two HeNe and Ar ion lasers, and emission filters (LP 560 nm for rhodamine, BP 505–530 nm for GFP, and BP 530–600 nm for yellow fluorescent protein).
Microarray analysis
For the determination of gene transcript levels, Col-0 and atmyb56#1 plants grown on 3% sucrose media were determined with microarray analysis. Ten-day-old Col-0 and atmyb56#1 mutant seedlings were frozen in liquid nitrogen before pulverization. Total RNA extraction was conducted using the RNeasy Plant Mini Kit (USA). RNA analysis was performed using Arabidopsis (V4) Gene Expression Microar-ray following the manufacturer’s instructions (Design ID: 21169, Agilent Technologies, USA).
Quantitative real-time PCR (qRT-PCR)
To conduct RT-PCR, total RNA samples were subjected to the reverse transcription reaction using M-MLV Reverse Transcriptase (Intron) with RNase inhibitor (Intron). Then, the cDNA was utilized for the qRT-PCR (cycling conditions:initial denaturation at 95°C for 10 min, followed by 42 cycles at 95°C for 15 s, 60°C for 1 min, and 95°C for 10 s, and a melt curve at 65°C to 95°C for 5 s; Nguyen et al., 2016). ACTIN2 represents as an internal control and the primers used in the PCR amplification are given in Supplementary Table S1.
RNA gel blot assay
Aurintricarboxylic acid and lithium chloride methods were used to extract total RNA from the sample plants (Lee et al., 2002). DNA probes were labeled with [α-32P] dCTP and a random primer mix (Invitrogen™, Life Technologies GmbH). Membranes were washed two times using 1 × SSC/0.1% SDS and once with 0.1 × SSC/0.1% SDS at 42°C before autoradiography. Gene-specific probes were prepared via PCR amplification with the following primers: AtMyb56-F, ATG AAT CCA AAT CTC CTT GAG AAA; and AtMyb56 – R, GGA AGC TCC AAC TCC AAG AAA A.
Histochemical GUS assay
A GUS staining solution consisting of 0.5 mM potassium ferricyanide, 10 mM EDTA, 100 mM sodium phosphate buffer (pH 7.0), 0.5 mM potassium ferrocyanide, 1 mM X-Gluc, and 0.1% Triton X-100 was manufactured. Seedlings were soaked in the GUS solution at 37°C for 1 h and chlorophyll removed by subsequent incubation in 70% ethanol for 6 h. Seedlings were then observed and photographed under a Leica EZ4D microscope (Jefferson et al., 1987). To generate AtMyb56pro:GUS reporter lines, a 1-kb promoter region of AtMyb56 was amplified with the primers of AtMyb56 F (5′-CGT TAT GGA GAG AAC AGA AAG CTC-3′) and R (5′-GTT TCT GGG TTT AGG GAT TAA GG-3′), inserted into the TOPO vector using a pCR8/GW/TOPOR TA CloningR Kit vector (Invitrogen™), and then transferred into the pMDC162 vector (Curtis and Grossniklaus, 2003). DNA sequencing was analyzed to confirm the integrity of this construct prior to the transformation of Arabidopsis (Columbia ecotype) via the Agrobacterium tumefaciens-mediated floral dipping method (Clough and Bent, 1998).
Measurement of starch content
Starch content was analyzed as described before (McCleary et al., 1994). Fourteen-day-old Col-0 and atmyb56#2 seedlings grown for 24 h on 0 or 200 mM sucrose-treated media were used to measure starch.
Measurement of sugar content
Fourteen-day-old Col-0 and atmyb56#2 seedlings grown for 24 h on 0 or 200 mM sucrose-treated media were used to measure sugar contents. Each dried sample of 0.5 g was ground using a blender, was treated with 5% of 1.5 mM trichloroacetic acid and vortexed for 1 min. The supernatant was filtered (PTFE membrane, 13 mm, 0.2 μm; Advantec, USA) after centrifugation at 10,000 × g for 15 min. The soluble sugar content was measured by high performance liquid chromatography using Dionex Carbopac™ (PA-1 [4 × 250 mm], P/N 35391 Dionex PAD), with 150 mM NaOH as solvent A, and 150 mM NaOH and 600 mM sodium acetate as solvent B. The conditions of elution were 0–5 min, 0% B; 5–15 min, 10% B; 15–20 min, 100% B; 20–30 min, 0% B, and the flow speed was 1 ml min−1. Glucose, fructose, sucrose, and maltose were used as standards to quantify sugar contents.
Statistical analyses
Each experiment was repeated at least three times using 10–300 samples in each repetition. We utilized one-way analysis of variance and Tukey’s test at 95% confidence level for statistical analyses.
RESULTS
AtMyb56 is induced by sucrose in Arabidopsis
Sucrose is a key factor in the pathways of flavonoid and an-thocyanin biosynthesis (Teng et al., 2005). Therefore, the present study attempted to identify which Myb genes are induced in response to sucrose. Using this approach, one gene, AtMyb56, was retrieved. Increased levels of AtMyb56 transcript were observed in a time-dependent manner after incubation in 60 mM sucrose (Fig. 1A). In the absence of sucrose, the lowest level of AtMyb56 expression was observed; however, increased sucrose treatment time and concentration led to a concomitant increase in the expression of AtMyb56 as determined by qRT-PCR (Fig. 1B). Sucrose can cause osmotic stress. Therefore, we applied qRT-PCR to examine AtMyb56 expression in 10-day-old Col-0 seedlings in response to various abiotic stresses and hormones. As shown in Fig. 1C, these stresses did not have a dramatic effect on AtMyb56 expression.
Fig. 1. Expression of AtMyb56 in plants subjected to stress.
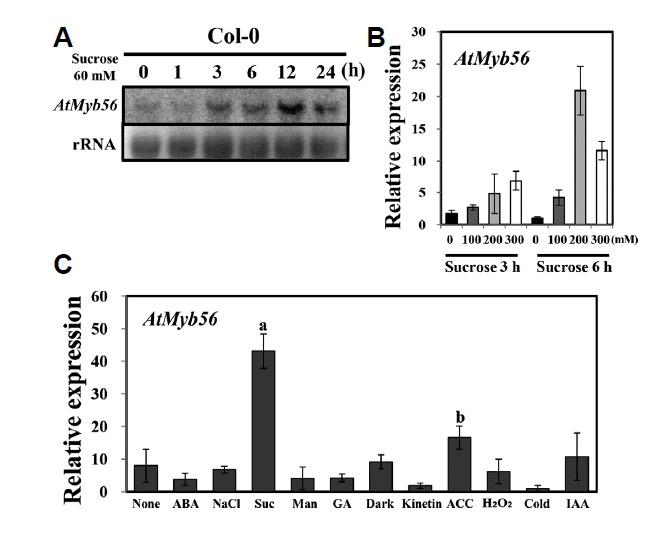
(A) Ten-day-old Col-0 plants exposed to 60 mM sucrose for 0, 1, 3, 6, 12, and 24 h. Total RNA is presented with a loading control. (B) Transcript levels of AtMyb56 in Col-0 seedlings exposed to different concentrations of sucrose were determined by qRT-PCR. Nine-day-old seedlings were treated with 0, 100, 200, or 300 mM sucrose for 3 or 6 h before total RNA was extracted. (C) Transcript levels of AtMyb56 in Col-0 seedlings exposed to different conditions were determined by qRT-PCR. Ten-day-old Col-0 plants were exposed to 10 μM ABA, 10 μM ethylene (ACC), 10 μM GA, 10 μM indole-3 acetic acid (IAA), 10 μM JA, 10 μM Kinetin, cold (4°C), 200 mM mannitol, 10 mM H2O2, 200 mM NaCl, 200 mM sucrose, and darkness for 3 h before total RNA was extracted. Error bars represent the standard errors (SEs) of three biological replicates. Values with different letters are significantly different from the Col-0 plants based on the analysis of variance (P <0.05).
The induction of anthocyanin accumulation by sugar, a well-known effective inducer, has been reported in many plant species including Arabidopsis seedlings. Since sucrose is not the only factor that can increase the accumulation of anthocyanin (Teng et al., 2005), the levels of AtMyb56 expression in the presence of other sugars were examined. Using a RNA gel blot assay, increased expression of AtMyb56 was detected following treatment with 200 mM glucose, maltose, or sucrose (Fig. 2A). This indicates that the expression of AtMyb56 is regulated by other sugars as well as sucrose. Next, we examined whether the AtMyb56 promoter gene was responsible for the responses toward these sugars. Transgenic seedlings harboring the GUS reporter gene driven by the AtMyb56 promoter (~1 kb) were exposed to glucose, maltose, and sucrose for 6 h. All these sugars increased GUS expression, with the highest expression observed in the presence of glucose (Fig. 2B).
Fig. 2. RNA gel blot hybridization of Col-0 wild type and GUS expression in AtMyb56pro:GUS lines in response to various sugars.
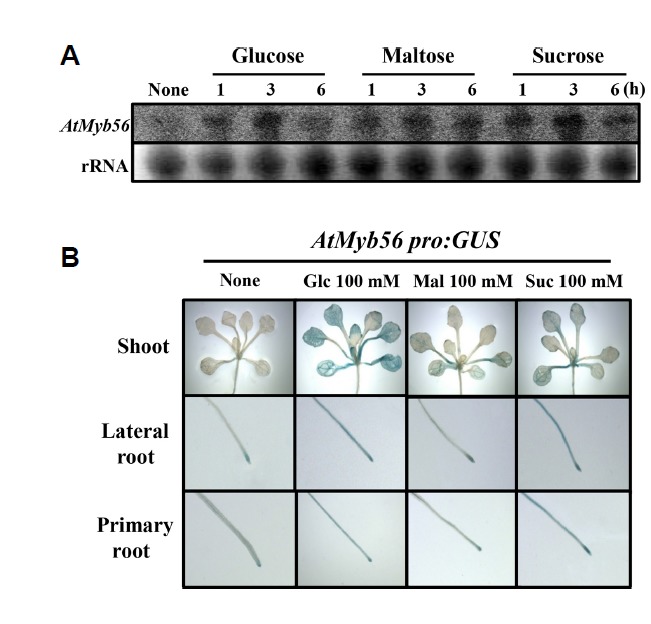
(A) RNA gel blot analysis of AtMyb56 gene expression following treatment with 200 mM glucose, maltose, or sucrose treatment for 0, 1, 3, and 6 h. Total RNA is presented with a loading control. (B) Representative images of Arabidopsis T3 homogenous transgenic seeds of AtMyb56pro:GUS lines under 100 mM glucose, maltose, or sucrose treatments for 3 h. Representative images are shown from an average of three independent experiments with 10 seedlings each.
AtMyb56 is localized to the nucleus
To verify the function of AtMyb56, T-DNA insertion mutants, SALK_060289 (atmyb56#1) and SALK_062413 (atmyb56#2), were obtained from the Arabidopsis Biological Resource Center (Fig. 3A). In addition, AtMyb56-overexpressing (AtMyb56 OE) lines were generated. RNA gel blot assays were used for confirmation of knockout or overexpressing (Fig. 3B). Usually, the nucleus localizes transcription factors. To investigate the nucleus localization of the AtMyb56 protein, a vector containing the full-length AtMyb56 cDNA and GFP gene was generated and transfected into onion epidermal cells (Fig. 3C). Fluorescence microscopy showed that GFP was uniformly distributed throughout the cells, whereas the AtMyb56-GFP fusion protein was localized exclusively in the nucleus (Fig. 3C). These results indicate that AtMyb56 protein expression is confined to the nucleus.
Fig. 3. Identification of atmyb56#1 and atmyb56#2 mutants and subcellular localization of AtMyb56 protein.
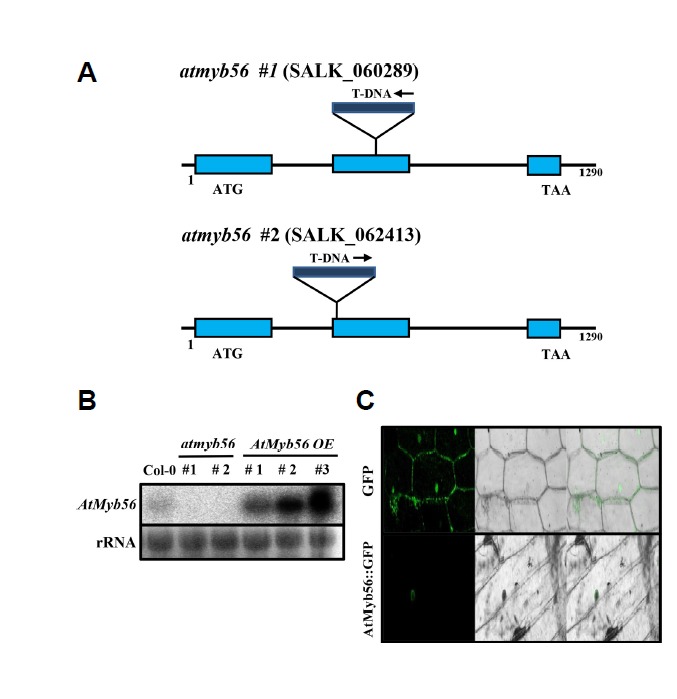
(A) Genomic structure of the AtMyb56 gene showing T-DNA insertions in mutants. (B) RNA gel blot analysis of AtMyb56 gene expression in atmyb56#1, #2, and AtMyb56 OE lines. Total RNA is presented with a loading control. (C) AtMyb56::GFP fluorescence in an onion epidermal cell.
Anthocyanin and flavonol levels are significantly decreased in atmyb56 mutants
Sucrose is important for anthocyanin and flavonol biosynthesis pathways. To investigate whether anthocyanin accumulation was controlled by AtMyb56, Col-0, atmyb56 mutant, and AtMyb56 OE seedlings were treated with sucrose. In the atmyb56 mutants, there was no significant increase in the accumulation of anthocyanin compared to in the Col-0 seedlings under normal conditions (Fig. 4A). Anthocyanin accumulation was strongly induced in Col-0 seedlings treated with 200 mM sucrose. However, atmyb56 seedlings were unable to accumulate anthocyanins to the same level as Col-0 seedlings in response to sucrose (Fig. 4A). Moreover, over-expressing of AtMyb56 increased the accumulation of an-thocyanin under normal condition (Fig. 4B). These results imply that sucrose-mediated anthocyanin accumulation was inhibited when the expression of the AtMyb56 gene was suppressed, which suggests that increased levels of sucrose stimulate the accumulation of anthocyanin in Col-0 seedlings, in part, via the regulation of AtMyb56. Furthermore, the distribution of flavonols in the roots of seedlings was examined and the DPBA fluorescence assay used to examine the level of flavonols accumulation in roots. In Fig. 4C, which shows the autofluorescence of DPBA staining in the root, brilliant yellow fluorescence corresponds to quercetin and green fluorescence to kaempferol. Based on the color intensity, the levels of both flavonols were found to be significantly decreased in atmyb56 mutants and increased in the AtMyb56 OE line. These results suggest that AtMyb56 positively regulates the accumulation of anthocyanins and flavo-nols.
Fig. 4. Anthocyanin and flavonol accumulation in Col-0, atmyb56 and AtMyb56 OE seedlings.
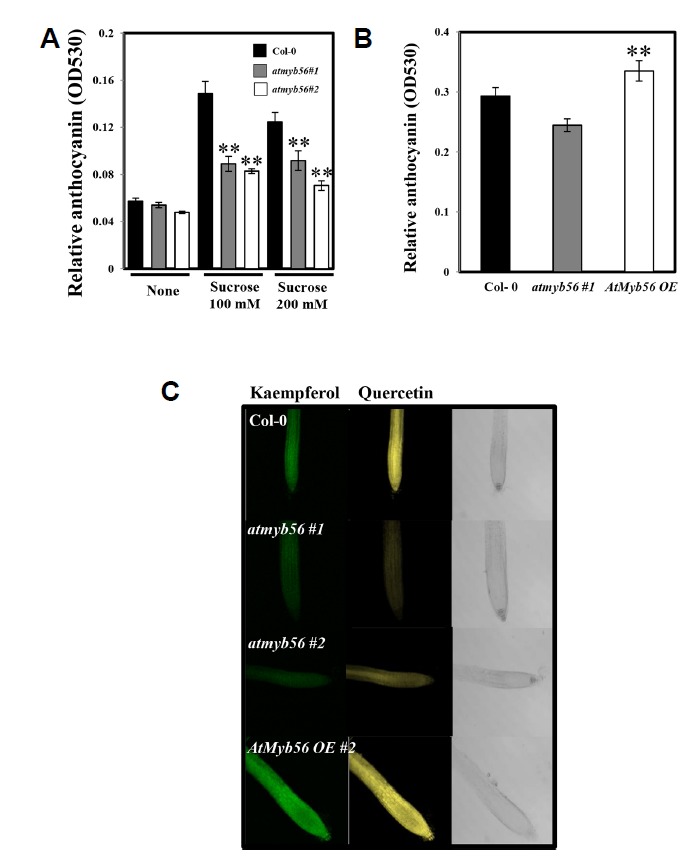
(A) Four-day-old seedlings of Col-0, atmyb56#1, and atmyb56#2 exposed to 0, 100 or 200 mM sucrose for 6 h were used for anthocyanin extraction. The anthocyanin content is shown, combined from two independent experiments with three replicates each. (B) Four-day-old seedlings of Col-0, atmyb56#1, and AtMyb56 OE#2 were used for anthocyanin extraction. The anthocyanin content is shown, combined from two independent experiments with three replicates each. (C) Roots of four-day-old Col-0, atmyb56#1, atmyb56#2, and AtMyb56 OE#2 seedlings were used for flavonol-DPBA staining to detect flavonol accumulation. Quercetin-DPBA conjugates display a yellow fluorescent color while kaempferol-DPBA conjugates show green fluorescence. Error bars represent the standard errors (SEs) of three biological replicates. Asterisks indicate significant differences from the Col-0 plants (P < 0.05).
AtGPT2 expression is altered in atmyb56 mutants
Since the accumulation of flavonoids in atmyb56 mutants is reduced and AtMyb56 is a Myb transcription factor, the expression of gene coding components of the anthocyanin biosynthesis was examined. The qRT-PCR was carried out for Col-0, atmyb56#1, and atmyb56#2 under different sucrose treatment conditions. As shown in Fig. 5, the expression levels of anthocyanin biosynthesis genes (PAP1, CHS, and FLS1) did not differ between mutants and Col-0 seedlings. In addition, the expressions of other anthocyanin biosynthesis genes, including CHI, DFR, and LDOX, were not significantly different in atmyb56 mutants and Col-0 seedlings (Supplementary Fig. S1). These results indicate that the decreased accumulation of anthocyanin in atmyb56 mutants might not be caused by the alteration of expression levels in gene coding components of the anthocyanin biosynthesis pathway.
Fig. 5. Determination of PAP1, CHS, and FLS1 transcript levels in Col-0, atmyb56#1, and atmyb56#2 seedlings exposed to sucrose.
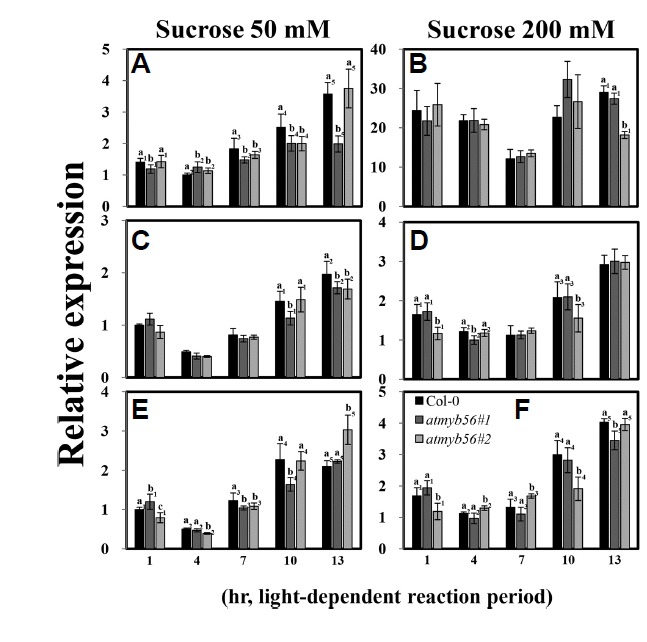
Transcript levels of PAP1(A and B), CHS (C and D), and FLS1 (E and F) in Col-0, atmyb56#1, and atmyb56#2 seedlings exposed to 50 and 200 mM sucrose were determined by qRT-PCR. Error bars represent the standard errors (SEs) of three biological replicates. Values with different letters are significantly different from the Col-0 plants based on the analysis of variance (P <0.05).
Since it is known that sugars enhance anthocyanin levels in plants (Larronde et al., 1998; Roubelakis-Angelakis and Kliewer, 1986), the levels of glucose, fructose, sucrose, and maltose were determined in Col-0 and atmyb56#2 mutant seedlings with or without sucrose treatment. There were no significant differences in levels of glucose, fructose, and sucrose between Col-0 and mutant seedlings. However, the atmyb56#2 seedlings were found to contain higher levels of maltose than did the Col-0 seedlings in the absence of sucrose (Fig. 6). In contrast, in the presence of 200 mM sucrose, atmyb56#2 seedlings exhibited decreased levels of maltose (Fig. 6).
Fig. 6. Sugar contents of Col-0 and atmyb56#2 seedlings with or without sucrose treatment.
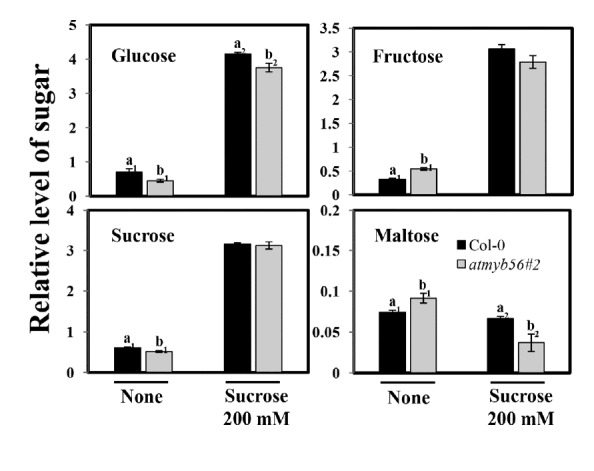
The sugar contents of 14-day-old Col-0 and atmyb56#2 seedlings grown for 24 h on media containing 0 or 200 mM sucrose were determined. Error bars represent the standard errors (SEs) of three biological replicates. Values with different letters are significantly different from the Col-0 plants based on the analysis of variance (P < 0.05).
Because it was difficult to predict which genes showed altered expression in the atmyb56 mutants, a preliminary mi-croarray analysis was utilized to compare gene expression between the mutant and Col-0 seedlings exposed to 3% sucrose (Table 1). In this analysis, several genes were found to have increased or decreased expression levels in the atmyb56 mutants. Among them, one candidate gene was identified, AtGPT2, which showed altered expression in the atmyb56 mutant. Plastidial glucose-6-phosphate/phosphate translocator (GPT) proteins mediate the transport of Glc6P from the cytosol to plastids leading to starch biosynthesis (Kammerer et al., 1998; Rolletschek et al., 2007; Zhang et al., 2008). Analysis of qRT-PCR showed that the transcript levels of AtGPT2 varied in Col-0 and atmyb56 mutants. Thus, sometimes AtGPT2 was more highly expressed in Col-0 than in atmyb56 mutants, and vice versa. AtGPT2 is thought to be involved in sugar transport and regulated by sugar levels or circadian rhythms (Kunz et al., 2010), which led to the transcript levels of AtGPT2 being examined over time and in response to sucrose in Col-0 seedlings and atmyb56 mutants. Col-0 seedlings and atmyb56 mutant samples were collected at four time points (1, 4, 7, and 10 h after light was turned on) in the presence or absence of sucrose (Fig. 7). As shown in Fig. 7A, in the absence of sucrose, the expression of AtGPT2 was low in the morning and gradually increased throughout the day in Col-0 seedlings. Similar patterns of AtGPT2 expression were detected when Col-0 seedlings were challenged with 50 and 200 mM sucrose. However, AtGPT2 transcript levels differed between the atmyb56 mutants and the Col-0 seedlings in the presence or absence of sucrose. As shown in Fig. 7A, in the absence of sucrose, AtGPT2 expression was higher in atmyb56 mutants than in Col-0 seedlings in the early time point after the light was turned on, but was lower in atmyb56 mutants than in Col-0 seedlings during the later time point after the light was turned on. However, when the seedlings were exposed to 50 mM sucrose, AtGPT2 expression was lower in atmyb56 mutants than in Col-0 seedlings throughout the entire period (Fig. 7B), while no significant differences were observed under the 200 mM sucrose treatment (Fig. 7C). These results suggest that AtMyb56 regulates the expression of AtGPT2 in response to circadian rhythms and sucrose.
Table 1.
Microarray analysis of differential gene expression in Col-0 and atmyb56 mutants. Transcripts exhibiting more than 2-fold difference between the wild-type and mutant plants are shown.
| MIPS | Description | Fold Change |
|---|---|---|
| Up – regulated gene | ||
| AT1G56650 | PAP 1 (PRODUCTION OF ANTHOCYANIN PIGMENT 1) | 3.2 |
| AT4G22880 | LDOX (LEUCOANTHOCYANIDIN DIOXYGENASE) | 6 |
| AT1G32900 | Starch synthase, putative | 7.2 |
| AT5G42800 | DFR (DIHYDROFLAVONOL 4-REDUCTASE) | 7.2 |
| AT1G60090 | BGLU4 (BETA GLUCOSIDASE 4) | 10.1 |
| AT4G03950 | Glucose-6-phosphate/phosphate translocator | 11.3 |
| AT5G61160 | AACT1 (Anthocyanin 5-aromatic acyltransferase 1) | 16.1 |
| AT4G15210 | BAM5 (BETA-AMYLASE 5) | 22.4 |
| AT1G56650 | GPT2 (Glucose-6-phosphate/phosphate translocator 2) | 78.5 |
| Down – regulated gene | ||
| AT2G43010 | PIF4 (phytochrome interacting factor 4) | −2.26 |
| AT1G28330 | DYL1 (DORMANCY-ASSOCIATED PROTEIN-LIKE 1) | −2.54 |
| AT1G76530 | Auxin efflux carrier family protein | −3.43 |
| AT2G33830 | Dormancy/auxin associated family protein | −3.74 |
| AT4G26530 | fructose-bisphosphate aldolase, putative | −4.09 |
| AT5G17800 | AtMyb56 | −36.67 |
Fig. 7. Determination AtGPT2 transcript levels in Col-0, atmyb56 #1, and atmyb56 #2 seedlings exposed to sucrose.
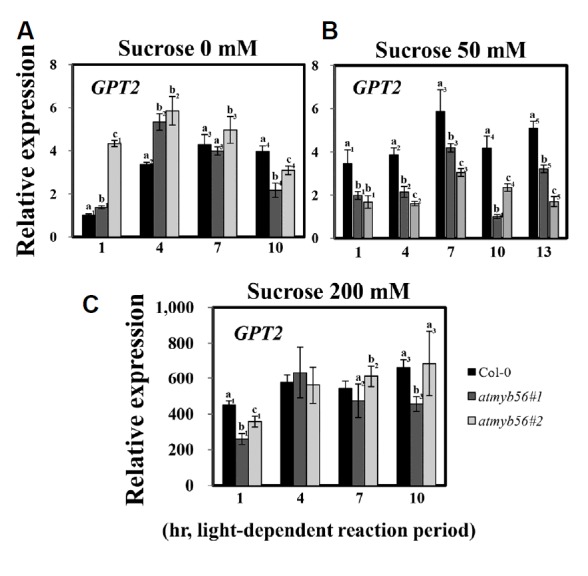
Transcript levels of AtGPT2 in Col-0, atmyb56#1, and atmyb56#2 seedlings exposed to 0 (A), 50 (B), and 200 mM sucrose (C) were determined by qRT-PCR. Error bars represent the standard errors (SEs) of three biological replicates. Values with different letters are significantly different from the Col-0 plants based on the analysis of variance (P < 0.05).
DISCUSSION
Previous studies have reported the crucial function of sucrose during anthocyanin accumulation in plants. Detached berries and grape cell suspensions have shown the enhancement of anthocyanin by sucrose (Larronde et al., 1998; Roubelakis-Angelakis and Kliewer, 1986). Moreover, sucrose has been shown to enhance the expression of MYB75/PAP1, which triggers the expression of anthocyanin biosynthesis genes (Solfanelli et al., 2006; Teng et al., 2005). The expressions of late anthocyanin biosynthesis genes including DFR, LDOX, UF3GT, and PAP1 are controlled by sucrose, whose products function downstream of the anthocyanin biosynthetic pathway, whereas its induction is lower in genes upstream of DFR (Solfanelli et al., 2006). Several hundred-fold changes of induction by sucrose are reported in these genes (Solfanelli et al., 2006). Sucrose acts as a signaling molecule during anthocyanin accumulation; however, the underlying signaling mechanisms, as well as the signaling components involved, remain largely unknown. It is of interest whether the effects of sugar on anthocyanin accumulation are adjusted by high levels of anthocyanin precursors; a recent study made some attempt to answer this question (Dai et al., 2014). These authors monitored variations in the level of the anthocyanin synthesis precursor, phenylalanine, in response to sucrose concentration. The higher the concentration of sucrose, the lower the concentration of phenylalanine detected, which indicated that the effects of sucrose were not due to the increased availability of the anthocyanin synthesis precursor (Dai et al., 2014).
To better understand the signaling cascade involved in sucrose-induced anthocyanin accumulation, a reverse genetics approach was adopted to identify Arabidopsis Myb genes involved in sucrose-induced anthocyanin accumulation. Myb genes were selected because previous studies have reported that various Myb transcription factors participate in flavonoid biosynthesis and in abiotic stress responses in plants (Shi and Xie, 2010; Stracke et al., 2001). In the first screening, AtMyb56, a sucrose-induced Myb gene, was identified as a R2R3 Myb family member in Arabidopsis. Accumulation of the AtMyb56 transcript increased in response to sucrose, reaching the highest level 6 h after exposure to 200 mM sucrose (Fig. 1B). Although, the expression of AtMyb56 was also observed in response to other signals such as ethylene, attention was focused on the sucrose response (Fig. 1C). In Arabidopsis, the efficiency of sucrose in anthocyanin accumulation was higher compared to glucose and fructose (Solfanelli et al., 2006). A similar induction of AtMyb56 was observed in the present study in response to glucose, maltose, and sucrose (Fig. 2). Significant changes in anthocyanin levels were detected under sucrose treatment, which resulted in an approximately two-fold decrease in anthocyanin in atmyb56 seedlings (Fig. 4A). In addition, a smaller decrease in flavonols (kaempferol and quercetin) was detected in atmyb56 seedlings (Fig. 4C). These results suggest that AtMyb56 and sucrose-induced anthocyanin accumulation had a positive correlation.
Since AtMyb56 contains a Myb domain, which can bind to DNA, possible candidate genes regulated by this protein were sought. First, genes involved in flavonoid biosynthesis were investigated, including PAP1, CHS, and FLS1. The transcript levels of these genes between Col-0 and atmyb56 seedlings in response to sucrose were not significantly different (Fig. 5). These results indicate that the decreased levels of anthocyanins in response to sucrose in atmyb56 seedlings were not caused by changes in the expression of flavonoid biosynthesis genes, which suggests that the decreased levels of anthocyanins in atmyb56 seedlings could be due to an indirect effect. Therefore, a preliminary microarray was employed to examine global changes in transcript levels in atmyb56 seedlings compared to WT seedlings. Microarray results obtained using Col-0 and atmyb56 seedlings exposed to 3% sucrose suggested that the expression of several genes associated with sucrose metabolism or transport may be altered in atmyb56 seedlings. Among the possible candidate genes, AtGPT2 was focused on because of its highest level of expression in atmyb56 seedlings. The plastidial phosphate antiporter family, which are located in the plastid membrane, are responsible for major fluxes of carbon in the non-photosynthetic plastids. Arabidopsis contains two glucose-6-phosphate (Glc6P) translocator genes (AtGPT1 and AtGPT2) (Kammerer et al., 1998; Knappe et al., 2003; Niewiadomski et al., 2005). The function of GPT is to transfer Glc6P into plastids where it can be utilized as a source of starch biosynthesis or in the oxidative pentose phosphate pathway (Kammerer et al., 1998; Rolletschek et al., 2007; Zhang et al., 2008). AtGPT1 has been found to play a crucial role in fatty acids synthesis in oilseeds and in the development of pollen and embryo sacs (Eastmond and Rawsthorne, 2000; Hutchings et al., 2005; Kang and Rawsthorne, 1994). Unlike AtGPT1, the homozygous knockout of AtGPT2 does not appear to be involved in the control of vegetative and generative development (Niewiadomski et al. 2005). AtGPT2 transcript levels differed in atmyb56 mutants compared to Col-0 seedlings in the presence or absence of sucrose (Fig. 7). In the absence of sucrose, AtGPT2 in atmyb56 mutants had greater transcript levels compared to Col-0 seedlings in the morning, but lower levels in atmyb56 mutants than in Col-0 seedlings in the evening (Fig. 7A). As there is a change of sugar levels depending on the difference in photosynthesis between day and night, we decided to determine whether the expression of AtGPT2 was regulated by circadian rhythm. It appeared that AtGPT2 was regulated by sucrose; however, it was not clear whether AtGPT2 expression was under the control of a circadian rhythm or not (Fig. 7). When the seedlings were exposed to 50 mM sucrose, AtGPT2 expression was lower in atmyb56 mutants than in Col-0 seedlings throughout the entire period (Fig. 7B), whereas no significant differences were observed in the 200 mM sucrose treatment (Fig. 7C). These results suggest that AtMyb56 regulates the expression of AtGPT2 in response to sucrose. Plant growth is regulated by metabolite levels, particularly to fluctuations of carbohydrates within the cell (Rolland et al., 2002; Solfanelli et al., 2006). Thus, it is plausible that the lack of AtMyb56 as well as fluctuations of carbohydrates affected AtGPT2 expression (Fig. 7).
It has been reported that both GPTs are functional Glc6P translocators; however, only AtGPT1 is essential because atgpt1 mutant seedlings showed severe defects in pollen development, whereas the loss of AtGPT2 function did not cause any defects in plant development (Niewiadomski et al., 2005). Therefore, sugar or starch levels are not dramatically altered in atmyb56#1 or atmyb56#2 seedlings compared to Col-0 seedlings (Fig. 6 and Supplementary Fig. S2). Nevertheless, maltose contents were distinctly different between Col-0 and atmyb56 seedlings (Fig. 6). This could be due to changes in the expression of AtGPT2 in atmyb56 seedlings. AtGPT2 appears to be regulated by environmental cues, which leads to subsequent adjustments in sugar levels. For example, the AtGPT2 gene was previously reported to be induced by exogenous sucrose (Knappe et al., 2003).
Sugars have pivotal roles in signal transduction and energy generation. Sugars are involved in hormone-like signal transduction as primary messengers throughout the life cycles of plants (Rolland et al., 2002; Rook and Bevan, 2003; Smeekens, 2000). Additionally, sugars stimulate growth and regulate nutrient mobilization, photosynthesis, and flavonoid biosynthesis (Koch, 1996; Rolland et al., 2002).
Taken together, these findings propose that altered AtGPT2 expression results in defects in sucrose metabolism in atmyb56 seedlings, resulting in altered levels of maltose. Thus, the decreased accumulation of anthocyanins observed in the atmyb56 seedlings was caused by changes in sugar metabolism because of altered AtGPT2 expression. This suggests that sucrose-induced AtMyb56 compromises the expression of AtGPT2 and the ability of the plant to control sugar transport under challenging environments.
Supplementary data
ACKNOWLEDGMENTS
The present study was supported by a grant from the National Research Foundation (to Hojoung Lee; grant #2017R1A2B4008706).
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 2000;12:2383–2394. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai ZW, Meddar M, Renaud C, Merlin I, Hilbert G, Delrot S, Gomès E. Long-term in vitro culture of grape berries and its application to assess the effects of sugar supply on anthocyanin accumulation. J Exp Bot. 2014;65:4665–4677. doi: 10.1093/jxb/ert489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Madhuvanthi R, Karthikeyan AS, Raghothama KG. Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol Plant. 2009;2:43–58. doi: 10.1093/mp/ssn081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias AP, Braun EL, McMullen MD, Grotewold E. Recently duplicated maize R2R3 Myb genes provide evidence for distinct mechanisms of evolutionary divergence after duplication. Plant Physiol. 2003;131:610–620. doi: 10.1104/pp.012047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Li S, An X, Liu X, Qin H, Wang D. Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J. Genet Genomics. 2009;36:17–29. doi: 10.1016/S1673-8527(09)60003-5. [DOI] [PubMed] [Google Scholar]
- Dos Reis SP, Lima AM, de Souza CR. Recent molecular advances on downstream plant responses to abiotic stress. Int J Mol Sci. 2012;13:8628–8647. doi: 10.3390/ijms13078628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dröge W. Free radicals in the physiological control of cell function. Physiol Rev. 2002;82:47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- Dubos C, Gourrierec JL, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul J, Alboresi A, Weisshaar B, Lepiniec L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 2008;55:940–953. doi: 10.1111/j.1365-313X.2008.03564.x. [DOI] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15:573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, Rawsthorne S. Coordinate changes in carbon partitioning and plastidial metabolism during the development of oilseed rape embryos. Plant Physiol. 2000;122:767–774. doi: 10.1104/pp.122.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. Regulation of the anthocyanin biosynthetic pathway by TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53:814–827. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- Hedbacker K, Carlson M. SNF1/AMPK pathways in yeast. Front Biosci. 2008;13:2408–2420. doi: 10.2741/2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchings D, Rawsthorne S, Emes MJ. Fatty acid synthesis and the oxidative pentose phosphate pathway in developing embryos of oilseed rape (Brassica napus L.) J Exp Bot. 2005;56:577–585. doi: 10.1093/jxb/eri046. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusion: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Martin C. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol. 1999;41:577–585. doi: 10.1023/a:1006319732410. [DOI] [PubMed] [Google Scholar]
- Kang F, Rawsthorne S. Starch and fatty acid synthesis in plastids from developing embryos of oilseed rape (Brassica napus L.) Plant J. 1994;6:795–805. [Google Scholar]
- Kammerer B, Fischer K, Hilpert B, Schubert S, Gutensohn M, Weber A, Flügge UI. Molecular characterization of a carbon transporter in plastids from heterotrophic tissues: the glucose 6-phosphate/phosphate antiporter. Plant Cell. 1998;10:105–112. doi: 10.1105/tpc.10.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodabande Z, Jafarian V, Sariri R. Antioxidant activity of Chelidonium majus extract at phenological stage. Appl Biol Chem. 2017;60:497–503. [Google Scholar]
- Knappe S, Flügge UI, Fischer K. Analysis of the plastidic phosphate translocator gene family in Arabidopsis and identification of new phosphate translocator-homologous transporters, classified by their putative substrate-binding site. Plant Physiol. 2003;131:1178–1190. doi: 10.1104/pp.016519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch KE. Carbohydrate-modulated gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:509–540. doi: 10.1146/annurev.arplant.47.1.509. [DOI] [PubMed] [Google Scholar]
- Kunz HH, Häusler RE, Fettke J, Herbst K, Niewiadomski P, Gierth M, Bell K, Steup M, Flügge UI, Schneider A. The role of plastidial glucose-6-phosphate/phosphate translocators in vegetative tissues of Arabidopsis thaliana mutants impaired in starch biosynthesis. Plant Biol (Stuttgart) 2010;12:115–128. doi: 10.1111/j.1438-8677.2010.00349.x. [DOI] [PubMed] [Google Scholar]
- Larronde F, Krisa S, Decendit A, Chèze C, Deffieux G, Mérillon M. Regulation of polyphenols production in Vitis vinifera cell suspension cultures by sugars. Plant Cell Rep. 1998;17:946–950. doi: 10.1007/s002990050515. [DOI] [PubMed] [Google Scholar]
- Lee H, Guo Y, Ohta M, Xiong L, Stevenson B, Zhu JK. LOS2, a genetic locus required for cold-responsive gene transcription encodes a bifunctional enolase. EMBO J. 2002;21:2692–2702. doi: 10.1093/emboj/21.11.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Jung YJ, Shin SY, Lee YH. The natural flavone eupatorin induces cell cycle arrest at the G2/M phase and apoptosis in HeLa cells. Appl Biol Chem. 2016;59(2):193–199. [Google Scholar]
- Li L, Ban ZJ, Li XH, Wu MY, Wang AL, Jiang YQ, Jiang YH. Differential expression of anthocyanin biosynthetic genes and transcription factor PcMYB10 in pears (Pyrus communis L.) PLoS One. 2012;7:e46070. doi: 10.1371/journal.pone.0046070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Piero AR, Puglisi I, Rapisarda P, Petrone G. Anthocyanins accumulation and related gene expression in red orange fruit induced by low temperature storage. J Agric Food Chem. 2005;53:9083–9088. doi: 10.1021/jf051609s. [DOI] [PubMed] [Google Scholar]
- Matus JT, Aquea F, Arce-Johnson P. Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol. 2008;8:83–98. doi: 10.1186/1471-2229-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleary BV, Solah V, Gibson TS. Quantitative measurement of total starch in cereal flours and products. J Cereal Sci. 1994;20:51–58. [Google Scholar]
- Nguyen NH, Jeong CY, Lee WJ, Lee H. Identification of a novel Arabidopsis mutant showing sensitivity to histone deacetylase inhibitors. Appl Biol Chem. 2016;59:855–860. [Google Scholar]
- Niewiadomski P, Knappe S, Geimer S, Fischer K, Schulz B, Unte US, Rosso MG, Ache P, Flügge UI, Schneider A. The Arabidopsis plastidic glucose 6-phosphate/phosphate translocator GPT1 is essential for pollen maturation and embryo sac development. Plant Cell. 2005;17:760–775. doi: 10.1105/tpc.104.029124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata K, Morikawa S, Nakamura H, Sekikawa A, Inoue T, Kanai H, Sarai A, Ishii S, Nishimura Y. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell. 1994;79:639–648. doi: 10.1016/0092-8674(94)90549-5. [DOI] [PubMed] [Google Scholar]
- Ohto MA, Onai K, Furukawa Y, Aoki E, Araki T, Nakamura K. Effects of sugar on vegetative development and floral transition in Arabidopsis. Plant Physiol. 2001;127:252–261. doi: 10.1104/pp.127.1.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polge C, Thomas M. SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci. 2007;12:20–28. doi: 10.1016/j.tplants.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Shuman J, Shulaev V, Davletova S, Mittler R. When defense pathways collide, the response of Arabidopsis to a combination of drought and heat stress. Plant Physiol. 2004;134:1683–1696. doi: 10.1104/pp.103.033431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F, Moore B, Sheen J. Sugar sensing and signaling in plants. Plant Cell (Suppl) 2002;14:S185–S205. doi: 10.1105/tpc.010455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolletschek H, Nguyen TH, Häusler RE, Rutten T, Göbel C, Feussner I, Radchuk R, Tewes A, Claus B, Klukas C, et al. Antisense inhibition of the plastidial glucose-6-phosphate/phosphate translocator in Vicia seeds shifts cellular differentiation and promotes protein storage. Plant J. 2007;51:468–484. doi: 10.1111/j.1365-313X.2007.03155.x. [DOI] [PubMed] [Google Scholar]
- Rook F, Bevan MW. Genetic approaches to understanding sugar response pathways. J Exp Bot. 2003;54:495–501. doi: 10.1093/jxb/erg054. [DOI] [PubMed] [Google Scholar]
- Rosinski JA, Atchley WR. Molecular evolution of the Myb family of transcription factors: evidence for polyphyletic origin. J Mol Evol. 1998;46:74–83. doi: 10.1007/pl00006285. [DOI] [PubMed] [Google Scholar]
- Ross JA, Kasum CM. Dietary flavonoids: bioavailability, metabolic effects, and safety. Annu Rev Nutr. 2002;22:19–34. doi: 10.1146/annurev.nutr.22.111401.144957. [DOI] [PubMed] [Google Scholar]
- Roubelakis-Angelakis KA, Kliewer WM. Effects of exogenous factors on phenylalanine ammonia-lyase activity and accumulation of anthocyanins and total phenolics in grape berries. Am J Enol Viticult. 1986;37:275–280. [Google Scholar]
- Seo PJ, Xiang F, Qiao M, Park JY, Lee YN, Kim SG, Lee YH, Park WJ, Park CM. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 2009;151:275–289. doi: 10.1104/pp.109.144220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi MZ, Xie DY. Features of anthocyanin biosynthesis in pap1-D and wild-type Arabidopsis thaliana plants grown in different light intensity and culture media conditions. Planta. 2010;231:1385–1400. doi: 10.1007/s00425-010-1142-9. [DOI] [PubMed] [Google Scholar]
- Smeekens S. Sugar-induced signal transduction in plants. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:49–81. doi: 10.1146/annurev.arplant.51.1.49. [DOI] [PubMed] [Google Scholar]
- Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P. Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol. 2006;140:637–646. doi: 10.1104/pp.105.072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol. 2001;4:447–456. doi: 10.1016/s1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S. Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 2005;139:1840–1852. doi: 10.1104/pp.105.066688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T, Nishiyama Y, Hirai MY. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 2005;43:218–235. doi: 10.1111/j.1365-313X.2005.02371.x. [DOI] [PubMed] [Google Scholar]
- Tsukaya H, Ohshima T, Naito S, Chino M, Komeda Y. Sugar-dependent expression of the CHS-A gene for chalcone synthase from petunia in transgenic Arabidopsis. Plant Physiol. 1991;97:1414–1421. doi: 10.1104/pp.97.4.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubi BE, Honda C, Bessho H, Kondo S, Wada M, Shozo K, Moriguchi T. Expression analysis of anthocyanin biosynthetic genes in apple skin: Effect of UV-B and temperature. Plant Sci. 2006;170:571–578. [Google Scholar]
- Wu HC, Jinn TL. Oscillation regulation of Ca2+/calmodulin and heat-stress related genes in response to heat stress in rice (Oryza sativa L.) Plant Signal Behav. 2012;7:1056–1057. doi: 10.4161/psb.21124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Shin Y. Antioxidant compounds and activities of edible roses (Rosa hybrida spp.) from different cultivars grown in Korea. Appl Biol Chem. 2017;60:129–136. [Google Scholar]
- Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, Zhigiang L, Yunfei Z, Xiaoxiao W, Xiaoming Q, et al. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol. 2006;60:107–124. doi: 10.1007/s11103-005-2910-y. [DOI] [PubMed] [Google Scholar]
- Zhang L, Häusler RE, Greiten C, Hajirezaei MR, Haferkamp I, Neuhaus HE, Flügge UI, Ludewig F. Overriding the co-limiting import of carbon and energy into tuber amyloplasts increases the starch content and yield of transgenic potato plants. Plant Biotechnol J. 2008;6:453–464. doi: 10.1111/j.1467-7652.2008.00332.x. [DOI] [PubMed] [Google Scholar]
- Zhang LC, Zhao GY, Jia JZ, Liu X, Kong XY. Molecular characterization of 60 isolated wheat MYB genes and analysis of their expression during abiotic stress. J Exp Bot. 2012;63:203–214. doi: 10.1093/jxb/err264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


