Fig. 2. Sequence alignment of SpLCD with LCDs and OCDs from other species.
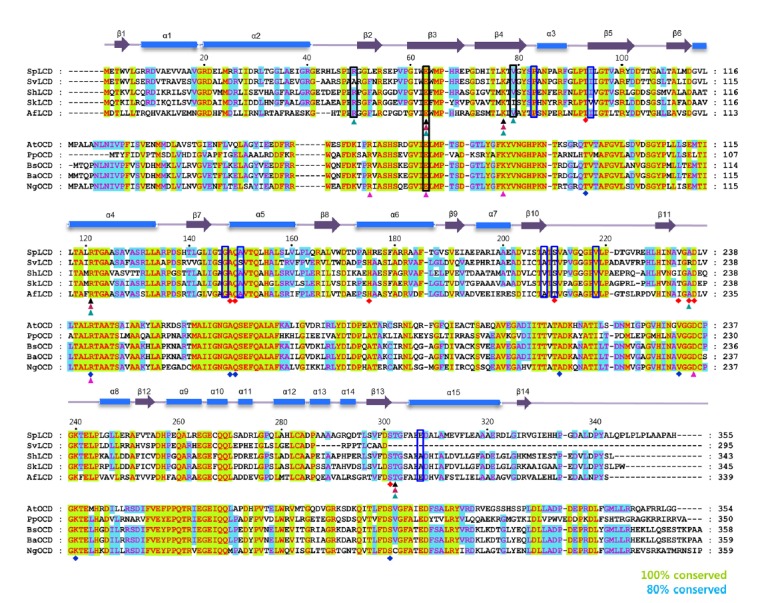
Multi-alignment of SpLCD (UniProtKB/Swiss-Prot accession numberD9UBW0) against LCD from Streptomyces virginiae(UniProtKB/Swiss-Prot accession numberQ8RR22), Streptomyces hygroscopicus(UniProtKB/Swiss-Prot accession numberA0A0A0NQJ4), Streptomyceskanamyceticus (UniProtKB/Swiss-Prot accession numberE9KTB3), Actinoplanes friuliensis(UniProtKB/Swiss-Prot accession numberU5VYY1), Agrobacterium tumefaciens (UniProtKB/Swiss-Prot accession numberQ9R468), Pseudomonas putida(UniProtKB/Swiss-Prot accession numberA0A179QU84), Brucella suis biovar 1(UniProtKB/Swiss-Prot accession numberA9WVR4), Brucella abortus biovar 1 (UniProtKB/Swiss-Prot accession numberC4IUX0), and Neorhizobium galegae by officinallis (UniProtKB/Swiss-Prot accession number A0A0T7GIJ3). Secondary structural elements were assigned using PyMOL and every twentieth residue is marked with a black dot. Strictly (100%) and semi-conserved residues (80%) are highlighted in green and light blue, respectively. Cylinders and arrows above the sequences denote α-helices and β-strands, respectively. Black, magenta, and cyan triangles below the LCD sequences denote L-pipecolic acid, L-proline, and L-DABA, respectively. Black boxes indicate residues interacting with water molecules when LCD or OCD bound its ligands and blue boxes denote residues interact with water molecules when LCD bound its cofactor; NAD+. Red and blue diamonds represent residues in the structures of SpLCD and Pseudomonas putida ornithine cyclodeaminase (PpOCD) that coordinate with NAD+ and NADH, respectively. Residues interacting with L-ornithine in OCD are shown in a pink triangle.
