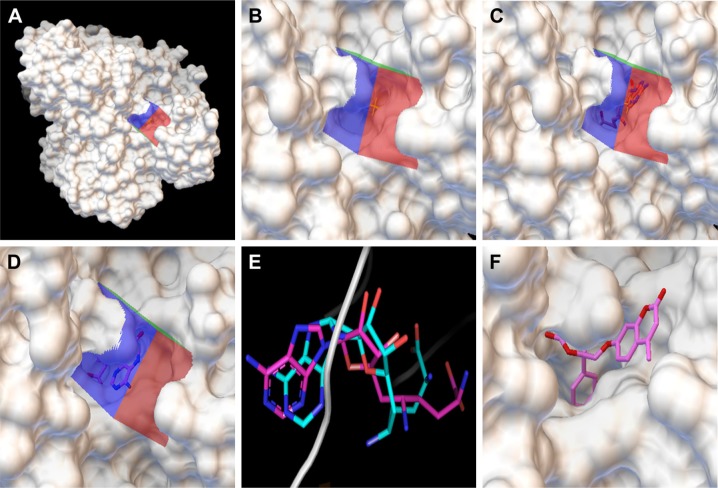
Figure 2.
Docking of compounds against the active site of BioA by employing virtual screening. (A) Virtual screening was performed by using Autodock4.2 with the substrate binding site of BioA as the target for docking. Surface representation of BioA structure is shown here with grid covering the active site. (B) Zoomed view showing the grid covering the active site of BioA. (C) Zoomed view of crystallographic binding of sinefungin binding at the active site of BioA. (D) Docking was performed with sinefungin and it was found to be localized in the grid. Zoomed view of docked sinefungin at the active site of BioA is represented here. Docking of sinefungin to the same site where sinefungin actually binds in the protein suggests the validity of the docking protocol. (E) Superimposition of crystallographic and docked modes of sinefungin at the active site of BioA, which displays almost the same binding pose. Pink color represents the crystallographic structure of sinefungin, and cyan color represents the docked structure of sinefungin. (F) Compound A36 docked at the active site by Autodock4.2. Zoomed view of the docked compound A36 is represented here. Color key: 1) white represents surface representation of proteins, 2) blue, red and green represent grid, 3) pink represents backbone of compound sinefungin and A36.
Abbreviation: BioA, 7,8-diaminopelargonic acid synthase.