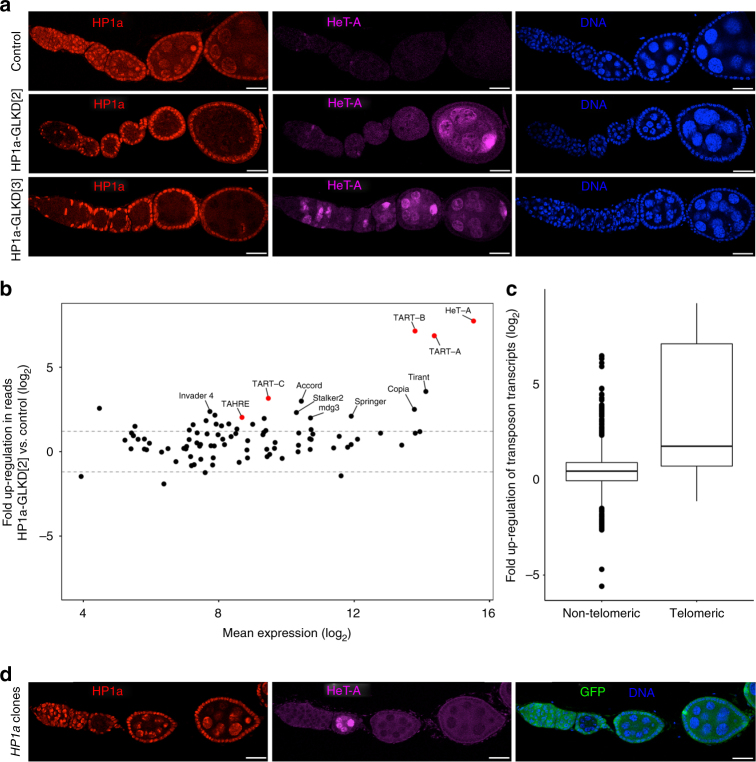
Fig. 1.
HP1a is required for the repression of selective transposons. a Representative images showing control and HP1a-GLKD fly ovaries stained for HP1a and HeT-A Gag proteins. Scale bars = 25 μm; n > 5. b Scatterplot showing the fold up-regulation of normalised RNA-seq reads mapping to canonical transposons in HP1a-GLKD compared to control ovaries. Telomeric transposons HeT-A, TART, and TAHRE are labelled in red. c Boxplot representing fold up-regulation in normalised RNA-seq reads mapping to telomeric and non-telomeric transposons in HP1a-GLKD vs. control. The middle line represents median. Box represents 25–75 percentile range called inter quartile range (IQR). Upper and lower whisker extend highest or lowest values till 1.5*IQR. Values above and below are outliers and plotted individually. d Ovaries with an HP1a mitotic clone (arrowhead) generated by FRT/FLP recombination stained for HP1a, GFP, and HeT-A Gag proteins. HP1a-null cells recapitulate show HeT-A upregulation. Scale bars = 25 μm; n = 3