Fig. 2.
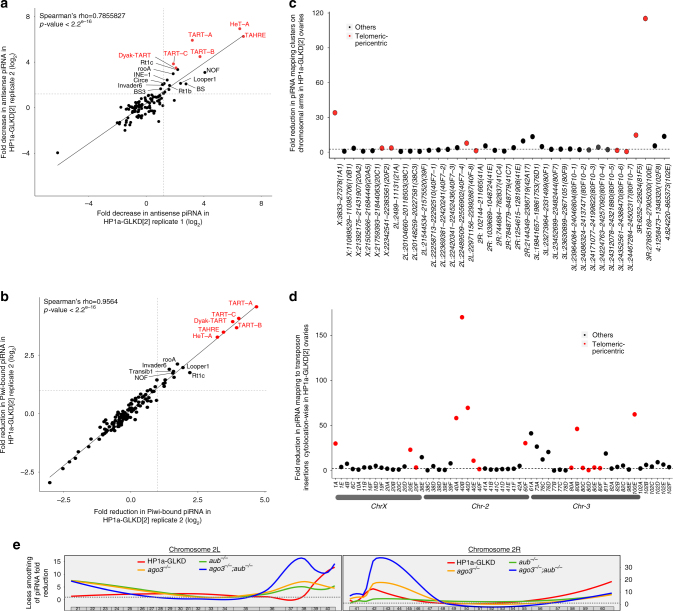
Loss of HP1a leads to the concomitant selective loss of piRNAs mapping to transposons. a, b Comparison of reduction in canonical transposon mapping piRNAs for a total cellular piRNAs in HP1a-GLKD[2] ovaries and b Piwi-bound piRNAs among two replicates of HP1a-GLKD[2] ovaries in comparison to control. Telomeric transposons, HeT-A, TART, and TAHRE, are labelled in red and correlation in piRNA loss among the replicates was estimated using Spearman’s rank correlation test. c Fold reduction in the total cellular piRNAs mapping to clusters on the chromosomal arms. Clusters close to the telomeres and centromeres are represented in red. d Fold reduction in the piRNAs uniquely mapping to transposon insertions, for different cytolocations. The piRNAs uniquely mapping to transposon insertions were consolidated for each cytolocation and the cytolocations having more than 1000 uniquely-mapping piRNAs in control were analysed. e Loess smoothening of the reduction in the 23–29-nt reads uniquely mapping to the transposon insertions in chromosome arms 2L and 2R. Fold reductions in those reads in HP1a-GLKD (HP1a-GLKD[2] and HP1a-GLKD[3] average), aub, ago3, and aub-ago3 ovaries compared with those in their respective controls are presented