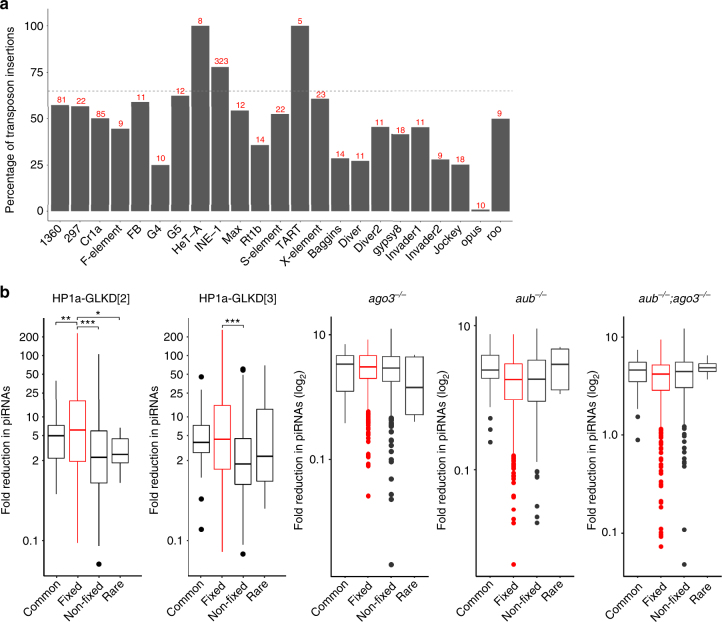
Fig. 5.
HP1a is required for the biogenesis of piRNAs mapping to evolutionarily older transposons. a The percentage of the transposons insertions from major transposon families that showed at least three-fold loss in the uniquely-mapping piRNAs in the HP1a-GLKD ovaries. The average reduction between the 23~29-nt read levels in the HP1a-GLKD[2] and HP1a-GLKD[3] ovaries was plotted, and the insertions targeted by at least 20 unique piRNAs were considered. The number in red represents number of such insertions for each transposon family. b Boxplots showing the reduction in the piRNAs mapping to the evolutionarily fixed, common, and rare transposon insertions, based on their occurrence in population, in the HP1a-GLKD and ago3, aub, and aub-ago3 double mutants. The significance of higher reduction of piRNAs mapping to evolutionarily fixed transposon insertions in comparison with those mapping to common or rare insertions was examined using Mann-Whitney U-test. The middle line represents median. Box represents 25–75 percentile range called inter quartile range (IQR). Upper and lower whisker extend highest or lowest values till 1.5 * IQR. Values above and below are outliers and plotted individually