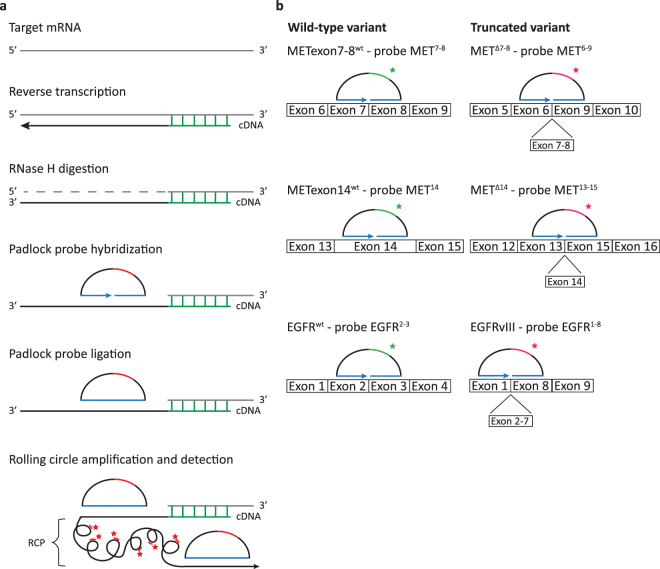
Figure 2.
In situ detection of whole-exon deletion splice variants using padlock probes. (a) Schematic overview of the method. mRNA is converted into localized cDNA molecules, which can be targeted through padlock probe hybridization and rolling circle amplification. Rolling circle products (RCPs) can then be detected with fluorescently labeled probes. Nucleic acid bridges in the BNA primer are depicted in green, padlock probe target sites are depicted in blue and padlock probe detection sites are depicted in red. (b) Design of padlock probes targeting METexon7–8wt, METΔ7-8, METexon14wt, METΔ14, EGFRwt and EGFRvIII. For specific detection of transcript splice variants, padlock probes were designed to target the mutation-specific exon-exon junction (detected with magenta fluorescence). For distinctive detection of their associated wild-type variants, padlock probes were targeted at the exon(s) missing in the exon deletion transcripts (detected with green fluorescence). Note that for graphical representation, the figure is not to scale.