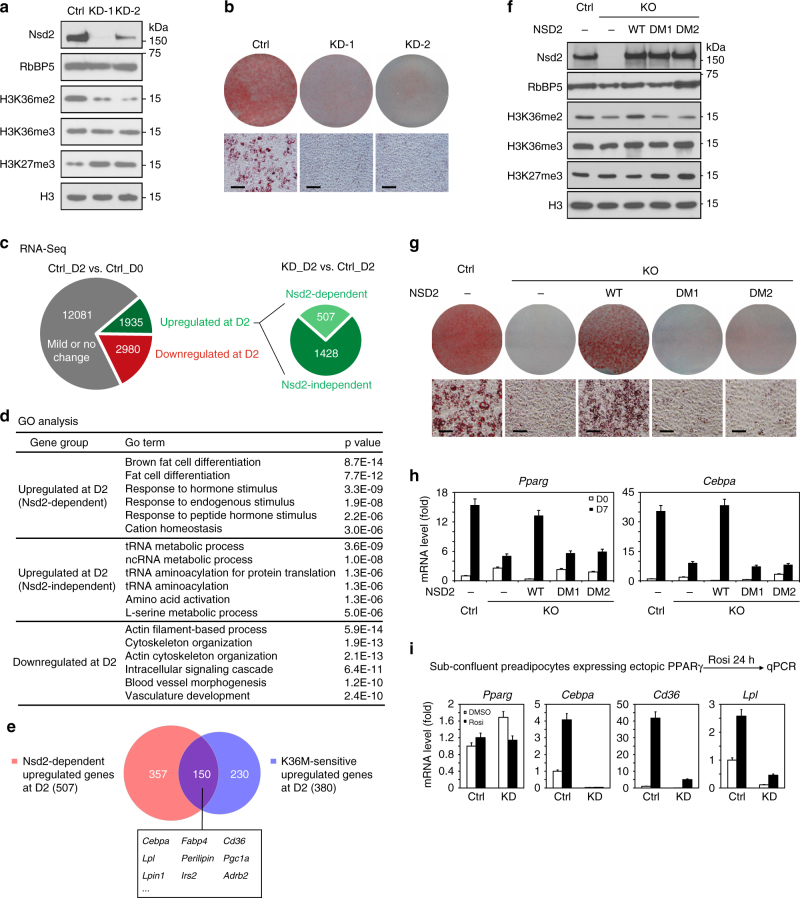
Fig. 3.
H3.3K36M targets H3K36 methyltransferase Nsd2 to inhibit adipogenesis and PPARγ target gene expression. a–e Immortalized brown preadipocytes were infected with lentiviral vector expressing control (Ctrl) or Nsd2 knockdown (KD) shRNAs, followed by adipogenesis assay until D7. Cells were collected at D0 and D2 for RNA-Seq. a Western blot of Nsd2 and histone methylations in preadipocytes. RbBP5 and histone H3 were used as loading controls. b Oil Red O staining at D7 of adipogenesis. Scale bars = 30 μm. c Schematic identification of Nsd2-dependent and Nsd2-independent up-regulated genes at D2 of adipogenesis. The threshold for up-regulation or down-regulation is twofold. d GO analysis of gene groups defined in c. e Venn diagram depicting Nsd2-dependent (507) and K36M-sensitive (380) up-regulated genes at D2. f–h Nsd2 methyltransferase activity is required for adipogenesis. Nsd2 knockout (KO) preadipocytes were generated using CRISPR. Control (Ctrl) and Nsd2 KO cells were infected with retroviral vector expressing either WT, Y1092A/Y1179A mutant (DM1), or H1042G/Y1179A mutant (DM2) human NSD2, followed by adipogenesis assay. f Western blot of Nsd2 and histone methylations in preadipocytes. g Cells were stained with Oil Red O at D7 of adipogenesis. Scale bars = 30 μm. h qRT-PCR of Pparg and Cebpa expression at D0 and D7 of adipogenesis. i Nsd2 is required for ligand-induced PPARγ target gene expression. Ctrl or Nsd2 KD preadipocytes were infected with retroviral vector expressing PPARγ. Sub-confluent cells were treated with DMSO or 0.5 μM Rosi for 24 h, followed by qRT-PCR of Pparg and its target genes Cebpa, Cd36, and Lpl. All qRT-PCR data are presented as means ± SEM. Three technical replicates from a single experiment were used