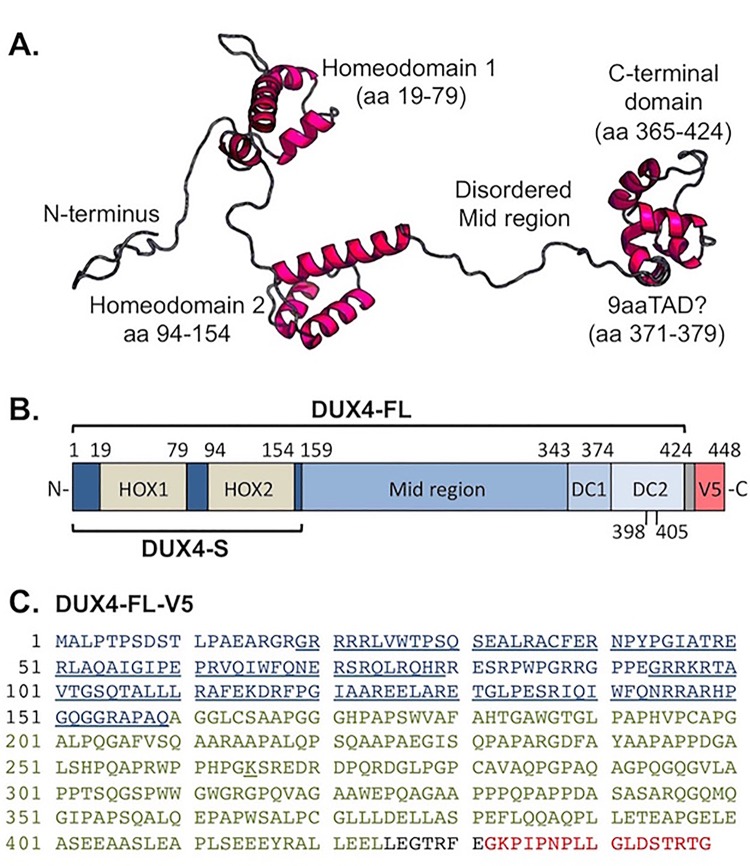
Fig. 1.
The DUX4 protein. (A) Ordered and disordered regions in the DUX4-FL protein as predicted by RaptorX Structure Prediction (raptorx.uchicago.edu). The two DNA-binding homeodomains and a C-terminal were predicted to have defined tertiary structures, whereas the ‘Mid’ region between homeodomain 2 and the C-terminal was predicted to be disordered. Shown is the most likely of the many similar structures returned by RaptorX. Similar predictions of ordered and disordered domains were generated by other prediction sites (not shown) as described in the Materials and Methods. In addition, there is a potential nine-amino acid transcription-activating domain (9aaTAD) at amino acids 371-379 as predicted by the online Nine Amino Acids Transactivation Domain Prediction Tool (http://www.med.muni.cz/9aaTAD/). (B) Linear representation of the DUX4 protein and sites of modification for this study. The diagram shows the two homeodomains, the predicted disordered Mid region, and sub-regions of the C-terminal domain as used to generate the DUX4 deletion and fusion cDNA constructs that are listed in Table 1. Each construct was modified by addition to the C-terminus of a seven-amino acid linker (gray unlabeled box) and the 17-amino acid V5 epitope. (C) Amino acid sequence of the full-length DUX4-FL-V5 protein as expressed in this study. The first 159 amino acids that compose the DUX4-S isoform are shown in blue with the two homeodomains underlined. The remaining amino acids (160-424) of endogenous DUX4-FL are shown in green, the linker sequence is in black, and the V5 epitope is in red.