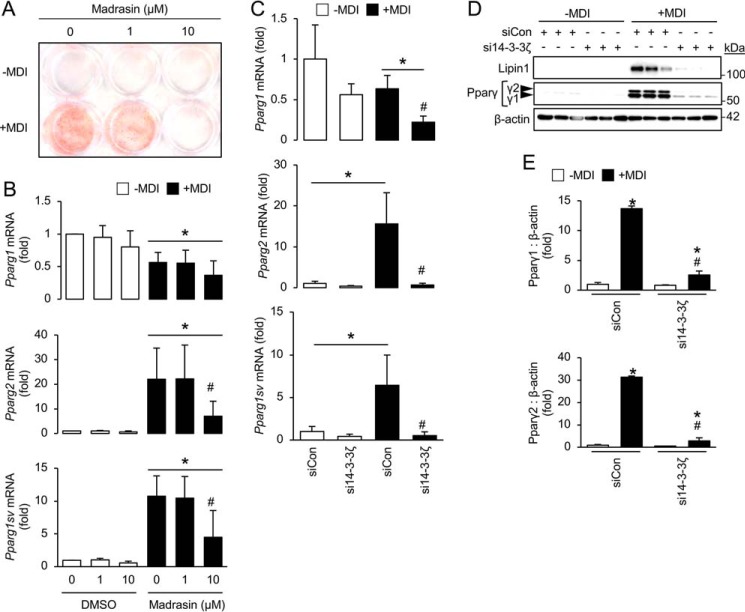
Figure 3.
Inhibition of the spliceosome or depletion of 14-3-3ζ prevents the alternative splicing of Pparg mRNA. A, 3T3-L1 cells were incubated with 1 or 10 μm madrasin in the presence of the adipogenic differentiation mixture (MDI), followed by differentiation for 7 days. Adipogenesis was assessed by Oil Red-O incorporation (representative of n = 5 independent experiments). B, RNA was isolated from madrasin-treated cells induced to differentiate for 48 h, and quantitative PCR was used to measure Pparg splice variants (n = 5 per group; *, p < 0.05 when compared with undifferentiated cells; #, p < 0.05 when compared with 0 μm madrasin, differentiated cells; bar graphs represent means ± S.D.). C, 3T3-L1 cells were transfected with siRNA against 14-3-3ζ or siCon and differentiated for 48 h, followed by isolation of total RNA to measure Pparg splice variants by quantitative PCR (n = 4 per group; *, p < 0.05 when compared with undifferentiated siCon-transfected cells; #, p < 0.05 when compared with differentiated, siCon-transfected cells; bar graphs represent means ± S.D.). D and E, 3T3-L1 cells were transfected with siRNA against 14-3-3ζ or siCon and differentiated for up to 7 days, followed by isolation of protein to measure Pparγ isoforms by immunoblotting (D). Protein abundance for each Pparγ isoform was measured by densitometry (E) (n = 4 per group; *, p < 0.05 when compared with undifferentiated siCon-transfected cells; #, p < 0.05 when compared with differentiated, siCon-transfected cells; bar graphs represent means ± S.D.).