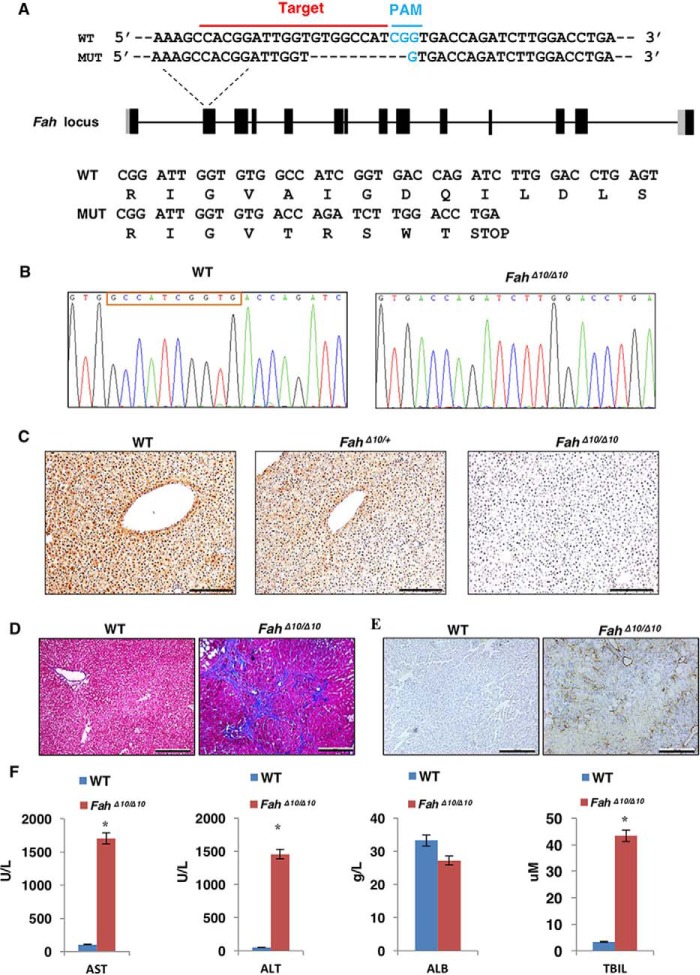
Figure 2.
Generation of the FahΔ10/Δ10 at and phenotype identification. A, the FahΔ10/Δ10 rat was generated by the CRISPR/Cas9 technology. sgRNA targeting the wildtype (WT) Fah sequence is indicated by the red line and the PAM sequence is labeled in blue. The reading frames of the WT and mutant Fah sequence are listed below. The 10-bp deletion in Fah results in an early STOP codon. B, sequencing results of the targeted Fah sites of WT and FahΔ10/Δ10 rats. The red box indicates the 10 missing bp in the FahΔ10/Δ10 rat. C, IHC staining to check Fah expression in WT, FahΔ10/+ and FahΔ10/Δ10 rat liver tissues. I, 100 μm. D and E, knockout of the Fah gene in rat caused severe liver damage including liver fibrosis/cirrhosis indicated by Masson's trichrome staining (D) and α-SMA (α-smooth muscle actin) (E) staining. F, WT and Fah rat serum AST, ALT, ALB, and TBIL levels were tested in WT and FahΔ10/Δ10 rats. *, p < 0.01 (n = 3) using two tailed unpaired Student's t test. Data are presented as mean ± S.D.