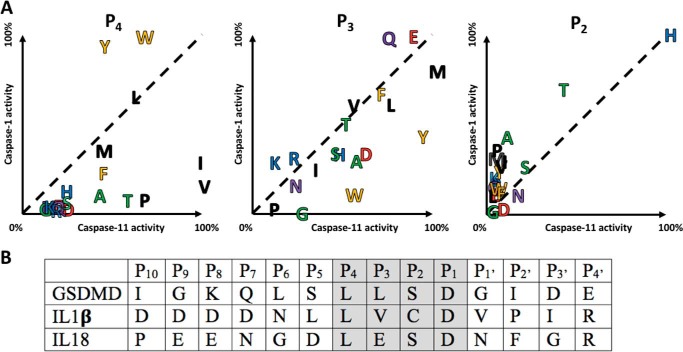
Figure 2.
Comparative subsite preferences. A, mouse caspase-1 and caspase-11 were screened on a natural amino acid positional scanning library, see Figs. S1–S3 for complete data. Relative activities reveal distinctive preferences at the P4 and P3 positions, and a common preference at the P2 position. There appeared to be no positions that could be used to discriminate caspase-1 from caspase-11 (except possibly Lys in the P3 position), but caspase-11 showed a preference over caspase-1 for β-branched residues in P4 and large hydrophobic residues in P3. Amino acids close to the dotted diagonal line are equally tolerated by both caspases. Residue coloring reflects common properties of the amino acid side chains. B, alignment of endogenous mouse substrates of the inflammatory caspases from the P10–P4′ region, with the region corresponding to synthetic substrates shaded.