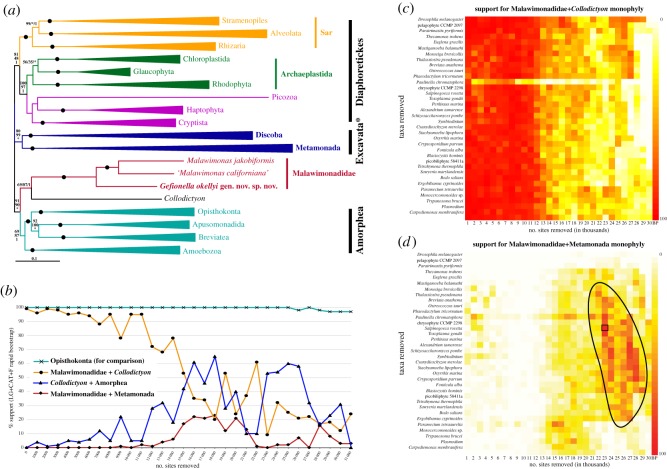
Figure 4.
(a) Phylogenetic tree of eukaryotes, based on 159 genes, with all sites and 84 taxa included. Maximum-likelihood (ML) tree shown was inferred under LG+C60+Γ4+F model of sequence evolution using IQ-TREE. For clades summarized as triangles, length shows average total branch length. ‘Excavata’ is labelled with an asterisk to signify that this clade does not include the ‘excavate taxon’ Malawimonadidae, and to also flag the contested nature of this clade (compare with figure 5). Statistical support values are, in order: LG+C60+Γ4+F model ‘ultrafast’ bootstrap approximation (UFboot) from IQ-TREE, LG+Γ4+F model bootstrap support (BP) from RAxML, and CAT-GTR+Γ4 model Bayesian posterior probabilities (from the two converged chains) in PhyloBayes-MPI. Filled circles represent maximal support (i.e. 100/100/1.0). Unlabelled branches received less than 50% UFboot support. Asterisks denote branches that were not recovered in inferred phylogeny for a given analysis. The full phylogenetic tree is shown in the electronic supplementary material, figure S5. (b) Chart depicting ML BP for groups on intervals along removal of fast-evolving sites. Y-axis denotes BP and X-axis denotes number of fastest-evolving sites removed. Support for Opisthokonta serves as an indicator of whether sufficient data is left for deep-level phylogenetic inference; other taxa summarized as triangles in a were similarly supported (data not shown). (c,d) Heat maps showing BP for a Malawimonadidae+Collodictyon clade (c) or a Malawimonadidae+Metamonada clade (d) with respect to removal of fast-evolving sites (X-axis, in thousands) and fast-evolving taxa (Y-axis). Pure white denotes 0% BP and pure red denotes 100% BP (rightmost columns demonstrate colour scale). Black box in d identifies dataset selected for full analysis shown in figure 5. Anomalous support values observed with removal of Paulinella chromatophora are probably result of unbalanced taxon sampling within Sar, which has an effect on overall tree topology. All analyses in b–d used LG+CAT+F model and rapid bootstrapping in RAxML.