Fig. 1.
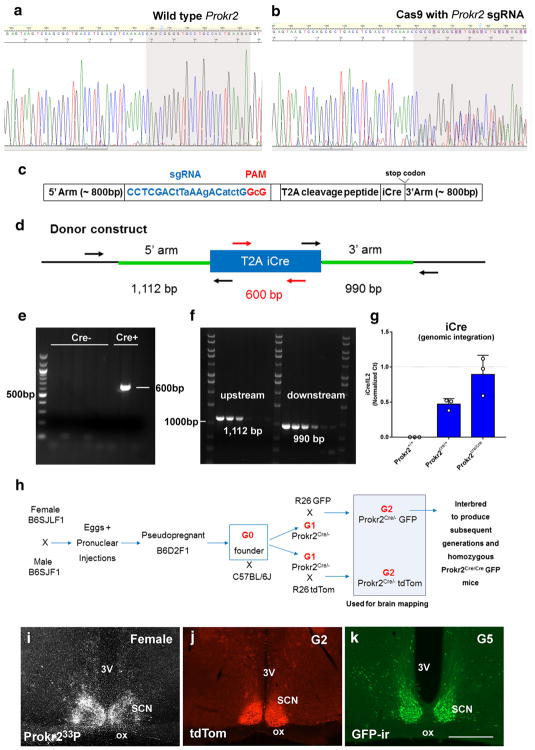
Generation of the Prokr2-Cre mouse line. a, b Chromatogram of DNA sequence from PCR products obtained by amplification across the Cas9 target shows the presence of indels in the genomic DNA due to non-homologous end joining repair of double-strand chromosome breaks induced by sgRNA/Cas9 complexes (b). c, d Schematic diagrams of the Prokr2-Cre donor construct used in the generation of Prokr2-Cre mouse model (c) showing the position of primers used to identify Prokr2-Cre founders (d). Note the position of the primers outside the homology arms to control for the correct insertion of Cre into the genome. e, f Images of agarose gels showing genotyping data of potential founders for the integration of Cre (e) and correct insertion of Cre sequence downstream of Prokr2 gene (different bands temperature gradient, f). g Bar graphs showing quantification analysis of iCre integration events using Il2 gene as a random positive control. Note that iCre and Il2 are found in the same dosage in Prokr2Cre/Cre mice, and iCre is reduced to half the amount of Il2 in Prokr2Cre/+ mice. h Schematic illustration of the breeding strategy used to obtain Prokr2-Cre reporter mice and Prokr2-Cre homozygous mice. i–k Digital images showing the distribution of Prokr2 mRNA (wild type mouse), Prokr2-Cre tdTomato and Prokr2-Cre GFP in the suprachiasmatic nucleus (SCN) in female brains (P60–80 days of age). 3V third ventricle, G2 generation 2, G5 generation 5, ox optic chiasm. Scale bar 200 μm