Abstract
As one of the world’s most important food crops, the potato (Solanum tuberosum L.) has spurred innovation in autotetraploid genetics, including in the use of SNP arrays to determine allele dosage at thousands of markers. By combining genotype and pedigree information with phenotype data for economically important traits, the objectives of this study were to (1) partition the genetic variance into additive vs. nonadditive components, and (2) determine the accuracy of genome-wide prediction. Between 2012 and 2017, a training population of 571 clones was evaluated for total yield, specific gravity, and chip fry color. Genomic covariance matrices for additive (G), digenic dominant (D), and additive × additive epistatic (G#G) effects were calculated using 3895 markers, and the numerator relationship matrix (A) was calculated from a 13-generation pedigree. Based on model fit and prediction accuracy, mixed model analysis with G was superior to A for yield and fry color but not specific gravity. The amount of additive genetic variance captured by markers was 20% of the total genetic variance for specific gravity, compared to 45% for yield and fry color. Within the training population, including nonadditive effects improved accuracy and/or bias for all three traits when predicting total genotypic value. When six F1 populations were used for validation, prediction accuracy ranged from 0.06 to 0.63 and was consistently lower (0.13 on average) without allele dosage information. We conclude that genome-wide prediction is feasible in potato and that it will improve selection for breeding value given the substantial amount of nonadditive genetic variance in elite germplasm.
Keywords: tetraploid, nonadditive effects, genome-wide prediction, potato, GenPred, Shared Data Resources, Genomic Selection
CULTIVATED potato (Solanum tuberosum L.) is unique among the major, global food crops in that it is autotetraploid and clonally propagated. As of 2018, there are 12 public breeding programs in the U.S. with a mandate to release varieties for commercial production, as well as several additional programs with a focus on germplasm enhancement. The variety development process begins with botanical seed from the sexual hybridization of heterozygous clones. Seedlings are grown in a greenhouse and one or more tubers from each plant are retained for subsequent vegetative propagation. Crossing and seedling tuber production takes 1–2 years—depending on the breeding program—followed by 1–2 years of field selection based primarily on visual assessment of tuber appearance, plant maturity, and yield components (tuber number and size), with some postharvest evaluation of processing traits such as fry color and specific gravity. Quantitative measurement of these traits in replicated and/or multi-location trials begins in field year (FY) 3 or 4 and continues for several years. Because it takes 3–4 years to establish clones as disease-free plantlets in vitro and to produce foundation seed, new varieties are typically released 10–12 years after crossing, by which time dozens of traits have been evaluated. The duration of the potato breeding cycle—from sexual hybridization to incorporating new clones as parents—is shorter than the time to variety release, but it is still 5–7 years for most U.S. programs. Until now, U.S. breeding programs have primarily used phenotypic selection in combination with genetic markers for a handful of major resistance genes (Lopez-Pardo et al. 2013).
The use of phenotype means for parent selection is not ideal because the estimates contain additive and nonadditive genetic effects, but only the former are efficiently transmitted to offspring (Gallais 2003). A number of previous studies have investigated nonadditive genetic variance in potato using factorial mating designs to estimate general and specific combining abilities (Plaisted et al. 1962; Tai 1976; Brown and Caligari 1989; Maris 1989; Neele et al. 1991; Gopal 1998; Bradshaw et al. 2000). General combining ability (GCA) is equivalent to the covariance between half-sibs, which equals for autotetraploid loci in panmictic and linkage equilibrium (Kempthorne 1955c; Gallais 2003). The symbols Va, Vd, and Vaa are the genetic variances for additive, digenic dominance, and additive × additive epistasis effects, respectively. Specific combining ability (SCA) is the covariance between full-sibs minus twice the covariance between half-sibs, which leads to an expression containing only nonadditive genetic variances The SCA/GCA ratio therefore provides an indication of the importance of nonadditive genetic variance. Although a wide range of values for SCA/GCA is found in the aforementioned references for potato, the general conclusion is that nonadditive genetic variance is important in many contexts.
For pedigreed populations, an alternative approach to estimating additive genetic variance is via the numerator relationship, or A, matrix. Kerr et al. (2012) were the first to publish a complete, recursive algorithm for A in autotetraploids, which has been applied to potato (Slater et al. 2014) and blueberry (Amadeu et al. 2016) populations. Mixed model analysis with A also enables selection on additive values calculated by BLUP (Henderson 1975). Although pedigree-BLUP is the cornerstone of genetic improvement for quantitative traits, the method has several limitations: (1) it depends on accurate pedigree records; (2) it neglects genetic covariance between founders; and (3) the covariance is based on expected, rather than realized, parental contribution.
Genomic selection (GS) has the potential to overcome these limitations by replacing A with a genomic covariance, or G, matrix estimated from markers (Bernardo 1994; Nejati-Javaremi et al. 1997; Habier et al. 2007; VanRaden 2008) or by estimating marker effects directly (Meuwissen et al. 2001). There have been several studies on GS in autotetraploid species (Annicchiarico et al. 2015; Li et al. 2015; Slater et al. 2016; Habyarimana et al. 2017; Sverrisdóttir et al. 2017), but none have used nonadditive genomic covariance matrices to partition genetic variance or predict total genotypic value. Both topics are addressed in this manuscript, building on analogous studies at the diploid level (Su et al. 2012; Vitezica et al. 2013; Xu 2013; Muñoz et al. 2014; Jiang and Reif 2015) and the classical theory of average effects in tetraploids.
Theory
In a series of articles in 1955 (Kempthorne 1955a,b,c), which were further distilled in the monograph An Introduction to Genetic Statistics (Kempthorne 1957), Kempthorne developed the theory of average effects for arbitrary ploidy, drawing upon the same mathematical methods used in the ANOVA for factorial experiments. Key results from this literature are reproduced here, as well as details on the derivation omitted by Kempthorne.
For an autotetraploid locus in panmictic equilibrium, assuming random bivalent formation (i.e., random chromosome segregation) and no inbreeding, the genotypic value (gijkl) of genotype ijkl (each index ranges from 1 to the number of alleles, and permutations of the indices are distinct) can be orthogonally decomposed into the population mean (μ) plus four additive effects (αi) corresponding to the four genes, six digenic dominance effects (βij) for all pairs of genes, four trigenic interactions (γijk), and one quadrigenic term (δijkl):
| (1) |
Equation 1 uses standard notation from the analysis of factorial experiments, where the symbols denote the regression coefficients and the regressors are implied to be indicator variables. Focusing on the additive effects and grouping the other parameters into a residual term, Equation 1 becomes a regression of genotypic value on allele dosage (Fisher 1941). The average effects minimize the sum of squared residuals for the population, which is equivalent to a sum over genotypes weighted by genotype frequency pijkl (= pipjpkpl under the assumptions of the model):
| (2) |
Taking the derivative with respect to the additive effect for each allele and equating the result to zero generates a set of normal equations that can be solved (Supplemental Material, Supporting Methods in File S1) to produce the following linear constraint:
| (3) |
which is identical to the result for diploids. By substituting Equation 3 into the normal equations (Equation S3, File S1), the solution for the additive effect of an allele becomes the average genotypic value of all individuals with that allele (multiple doses contribute separately), relative to the population mean:
| (4) |
The residuals from the regression equation for the additive effects, which we denote by are known as the dominance deviation. In diploids, this deviation uniquely defines one dominance effect for each genotype; but in tetraploids the dominance deviation comprises digenic, trigenic, and quadrigenic effects (Equation 1). The tetraploid solution for the digenic effects corresponds to regressing the dominance deviation on the dosage of pairs of alleles, which involves minimizing the following sum of squared residuals:
| (5) |
Taking the derivative with respect to the digenic effect for each allele pair and equating the result to zero generates a set of normal equations that can be solved (Supporting Methods, File S1) to produce the following linear constraint for any allele i:
| (6) |
which is the same result for diploids. Substituting Equation 6 into the normal equations (Equation S6, File S1) leads to the solution
| (7) |
By now the pattern is clear and the least-squares solution for the trigenic effects can be written as
| (8) |
Having solved for the additive, digenic, and trigenic terms; the quadrigenic effect δijkl is the residual in Equation 1.
The breeding value (BV) of an individual is defined as twice the mean genotypic value of its progeny relative to the population mean. Under the model assumptions, all six possible gene pairs for tetraploid genotype ijkl have equal frequency in its gametes which, in conjunction with Equation 7, leads to the following expression:
| (9) |
Equation 9 shows that breeding value equals the total additive value (u) plus 1/3 of the total digenic dominance (v), but it is conventional to refer to the additive value as “breeding value” because the contribution of digenic dominance diminishes exponentially: 1/3t after t generations (Gallais 2003). This is analogous to the situation in diploids (and polyploids) with regard to additive × additive epistasis, as it contributes to a breeding value with a coefficient of 1/2 but is generally omitted when referring to breeding value.
Materials and Methods
Training population
Phenotype data for a training population (TP) of 571 round white clones was collected between 2012 and 2017 at the University of Wisconsin (UW–Madison) Hancock Agricultural Research Station (number of clones trialed per year is in Table S1 in File S1). Between 2012 and 2015, all clones were entries in the National Chip Processing Trial (NCPT) which were contributed by 11 public U.S. breeding programs. In 2016 and 2017, FY3 and FY4 selections from the UW–Madison breeding program were included in addition to the NCPT clones. The NCPT uses a two-tier evaluation system, with one plot per location for tier 1 clones and two plots per location for tier 2 clones. FY3 clones were evaluated with a single plot, and FY4 clones were evaluated with two plots in 2016 and one plot in 2017. All plots contained 15 seed pieces, planted with 30-cm in-row spacing and 91 cm between rows. Trials were planted in late April and harvested in early September, with vine desiccation 2–3 weeks before harvest.
Phenotype data for three traits—yield, specific gravity, and fry color—are included in this study. Total yield is based on the weight of all harvested tubers and reported in Mg ha−1. Specific gravity was determined by water displacement, using 2–3 kg of tubers per plot (Wang et al. 2017). Fry color was measured in March of each year after 6 months of storage (1 month at 12.8° for wound healing, followed by 5 months at 8.9°), using 1-mm slices fried for 130 sec in vegetable oil at 182°. For the 2012–2014 trials, fry color was measured on a 1–10 visual scale, while for the 2015–2017 trials it was measured on the L* lightness scale using the D25 HunterLab Colorimeter (Hunter Associates Laboratory). Fry color measurements on the visual scale (x) were converted to L* using the formula L* = −1.37x + 63.7, which is based on a linear regression analysis of 70 clones phenotyped with both methods.
TP samples were genotyped with either version 1 or version 2 of the Solanaceae Coordinated Agricultural Project potato SNP array, which have in common a set of 8303 markers used for this study (Hamilton et al. 2011; Felcher et al. 2012). Tetraploid genotype calls (coded 0–4) were made using version 1.6 of the ClusterCall package (Schmitz Carley et al. 2017) in R (R Development Core Team 2015), which calibrates the relationship between signal intensity from the array and allele dosage for each marker based on multiple F1 populations. In addition to the Atlantic × Superior, Wauseon × Lenape, and Rio Grande × Premier Russet populations used by Schmitz Carley et al. (2017); two more calibration populations were used: Waneta × Pike (da Silva et al. 2017; n = 184) and A06084-1TE × Castle Russet (n = 245). Default parameters were used except for min.train = 3, which required a marker to be called in at least three of the five calibration families. The curated marker set contained 3895 polymorphic SNPs with ≥95% concordance across samples (File S2).
The numerator relationship matrix A was calculated with R package AGHmatrix (Amadeu et al. 2016) using pedigree records maintained by the authors as well as a public database (van Berloo et al. 2007). After removing uninformative ancestors, there were 185 founders (clones with no parent) and 1138 total clones in the pedigree (File S3).
Genomic covariance matrices
In the Theory section, average effects were derived for an arbitrary number of alleles. For biallelic SNPs, additional simplifications are possible. Consider alleles B and b with frequencies p and q, respectively. In this case, Equation 3 becomes which reduces to the following well-known formulas involving
| (10) |
If X denotes the dosage of B, then the total additive value is
| (11) |
where W is a centered genotype because 4p is the population mean of X. To obtain a similarly parsimonious expression for the total digenic dominance v, we follow the example of Wricke and Weber (1986) and introduce the parameter When combined with Equation 6, the result is (Supporting Methods, File S1)
| (12) |
| (13) |
For mixed model analysis, Gij quantifies the covariance between the additive values for clones i and j relative to the additive genetic variance
| (14) |
The covariance in Equation 14 involves the expectation with respect to but the additive genetic variance is based on the theory of average effects in which the expectation is with respect to genotypes. To relate the two variance components, the expectation with respect to both parameters is used:
| (15) |
Upon substituting Equation 15 into Equation 14 and extending the analysis to m loci in linkage equilibrium, the result is
| (16) |
The digenic dominance matrix Dij is defined similarly as the covariance between dominance values relative to the dominance genetic variance, based on the expectation with respect to
| (17) |
Using the following result for the dominance genetic variance
| (18) |
the D matrix is
| (19) |
For the covariance between additive × additive epistatic effects, we used the Hadamard product of the G matrix, denoted by G#G (Henderson 1985; Su et al. 2012).
Mixed model analysis of the TP data set
Stage-wise analysis of the multi-year TP data set was performed using ASReml-R version 3 (Butler et al. 2009) and a diagonal weight matrix to account for the varying precision of the estimates in the first stage (Smith et al. 2001; Damesa et al. 2017). Stage one was an analysis within a year, including blocking effects when present and modeling the genotype effect for each clone as fixed. The covariance matrix () for the vector of genotype effect estimates () in year j was obtained from the inverse of the coefficient matrix of the mixed model equations (Henderson 1975), which is returned as Cfixed in the asreml object. Stage two was a multi-year analysis based on the following linear model:
| (20) |
In Equation 20, the parameter φ is the intercept, gi is a random effect for genotype, yj is a fixed effect for year, (gy)ij is a random effect for the genotype × year interaction, and the variance of the residual fij is (ωij)−1, where ωij is the i-th diagonal element of from stage one (Damesa et al. 2017). File S4 contains the genotype effect estimates () and corresponding weights (ωij) used in the multi-year analysis.
After fitting a baseline model with independent genotype effects (var[gi] = Vg), five genetic models with different covariance structures for gi were tested (Table 1). The variance of the genotype × year interaction (var[gyij] = Vgy) was estimated in the baseline model and constrained at that value for the other models. This allowed for the partitioning of the genetic variance (Vg) into additive (Va) and residual (Vr) genetic components for models A and G. Models G+GG, G+D, and G+GG+D involved the estimation of nonadditive variance components for digenic dominance (Vd) and/or additive × additive epistasis (Vaa). Variances are reported using the standardization proposed by Legarra (2016) to make them comparable, in which the parameter estimate is multiplied by the difference between the mean of the diagonal elements and the mean of all elements of the covariance matrix Goodness of fit was assessed by the Akaike information criterion (AIC), defined as the deviance minus twice the number of variance parameters (Piepho 2009).
Table 1. Covariance structures for the total genotypic value (g) in Equation 20.
| Model | Var[g] |
|---|---|
| A | IVr + AVa |
| G | IVr + GVa |
| G+GG | IVr + GVa + (G#G) Vaa |
| G+D | IVr + GVa + DVd |
| G+GG+D | IVr + GVa + (G#G) Vaa + DVd |
g, total genotypic value.
Our objective was to compare how well the different covariance models predicted the total genotypic value g of unobserved clones. Each of the models in Table 1 has the form where θ is a sum of average effects and r is the residual genetic effect. Genomic predictions were calculated as from a stage-two analysis (Equation 20) without response values () for clones in the validation set, using the variance parameter estimates from ASReml-R and custom scripts to solve Henderson’s mixed model equations (Henderson 1975). The validation data were calculated as from a stage-two analysis with all clones, assuming independent clone effects (i.e., the baseline model). The reliability () of the validation data was calculated from the prediction error variance (PEV) by for clone i (Clark et al. 2012). To estimate prediction accuracy () from predictive ability (i.e., the Pearson correlation between the genomic predictions and validation data, ), the latter was divided by the square root of the mean reliability (i.e., broad-sense heritability) of the validation data (Dekkers 2007). Because the mean-squared error of the accuracy estimate worsens as the reliability of the validation data decreases (Ould Estaghvirou et al. 2013), only validation data with reliability ≥0.6 were used.
Genome-wide prediction in F1 populations
As part of various research projects, six unselected F1 populations (Table 2) were evaluated at the same location as the TP during the same time period. Populations W12011, W12012, and W12060 were evaluated for yield and specific gravity with a single plot of 12 plants per clone in 2015 and 2016. Populations W9817 and W10010 were evaluated for yield with two 8-plant plots in 2013 and one 20-plant plot in 2014 (Rak and Palta 2015). Population W×L was evaluated with a single plot for specific gravity for 4 years (2012–2015), but yield was only measured in 2014 and 2015; there were 6 plants per plot in 2012 and 10 plants per plot in 2013–2015 (Frederick 2017). Phenotype data were analyzed separately for each F1 population using a linear model with fixed effects for year and independent random effects for clone. Genetic and residual variance components were estimated with ASReml-R and used to calculate BLUPs for validation. The BLUPs and corresponding reliabilities (which were used to estimate accuracy from predictive ability, as described above) are provided in File S5; however, yield BLUPs for W12011 and W12060 were excluded because of low reliability (<0.6).
Table 2. Parentage, population size, and number of polymorphic markers in unselected F1 populations.
| Population | Mother (A90) | Father (A90) | < G90 > | Population size | No. markers |
|---|---|---|---|---|---|
| W12011 | W6360-1rus (0.01) | Silverton Russet (0.01) | 0.059 | 58 | 1580 |
| W12012 | W8736-6rus (0.06) | Silverton Russet (0.01) | 0.062 | 55 | 1744 |
| W12060 | Russet Norkotah (0.01) | Canela Russet (0.01) | 0.062 | 65 | 1376 |
| W9817 | Liberator (0.16) | W4013-1 (0.12) | 0.070 | 76 | 2311 |
| W10010 | Tundra (0.31) | Bannock Russet (0.01) | 0.066 | 48 | 1629 |
| W×L | Wauseon (0.20) | Lenape (0.25) | 0.070 | 167 | 1999 |
A90, 90th percentile of pedigree relationship with the training population; G90, 90th percentile of G coefficient between an F1 individual and the TP; <G90>, the average G90 for the F1 population.
The F1 populations were genotyped using the same SNP array as the TP. Tetraploid genotype calls (coded 0–4) were made in two ways: (1) as described above (using CC.anypop in the ClusterCall package) for the same set of 3895 markers selected for the TP, and (2) using the CC.bipop function. Polymorphic markers with identical calls for the two approaches were used for prediction (File S6). Genomic predictions for each F1 population were calculated as from a stage-two analysis of the entire TP (Equation 20), using the TP variance estimates with multi-population covariance matrices (“G_New” from Wientjes et al. 2017) to account for different allele frequencies in the TP vs. F1 population. If and denote scaled versions of the matrices defined previously:
| (21) |
then, using the subscripts 1 and 2 to denote the different populations, the covariance matrices are
| (22) |
Data availability
All marker, pedigree, and phenotype data needed to reproduce the results in this manuscript are provided in File S2, File S3, File S4, File S5, and File S6:
Results
Pedigree information was used to calculate the numerator relationship, or A, matrix for a TP of 571 clones. All but 13 clones had a pedigree depth (maximum number of generations from a founder) of at least 7, with a median depth of 10 generations (distribution in Figure S1 in File S1). For autotetraploids, diagonal element Aii is related to the inbreeding coefficient Fi—defined as the probability that two randomly chosen genes, sampled without replacement, are identical by descent—via the equation Aii = 1+3Fi (Gallais 2003). Values for Aii ranged from 1 to 1.55, with a mean of 1.05, so the most inbred clone in the population had Fi = 0.18.
In the G-BLUP method of GS, the covariance between additive values is proportional to a G matrix calculated from markers (instead of A). We calculated G using 3895 polymorphic markers from the potato SNP array for which accurate allele dosage information was available. The overall scaling of G is such that the mean of the diagonal elements equals 1 at panmictic equilibrium, which is very near the observed value of 0.99. As further confirmation, the observed frequency of heterozygotes was inspected as a function of allele frequency and found to be in close agreement with the expected values under panmixis (Figure S2 in File S1).
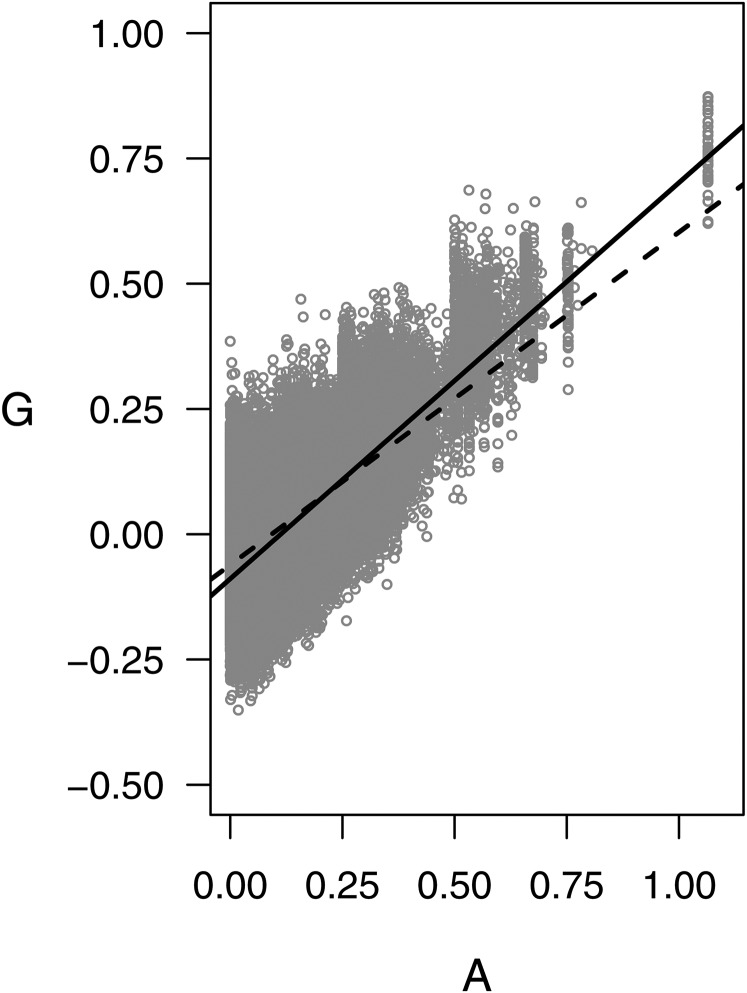
G has been called the “realized relationship matrix” because it captures Mendelian segregation around the expected value A (Hayes et al. 2009). This connection is the motivation for regression analysis between G and A (VanRaden 2008) and is shown in Figure 1 for our potato data set. The dashed line is the fit of the linear model when all off-diagonal elements are used (G = 0.66A–0.06, R2 = 0.41), which underestimated G at high values of A. By excluding very distant relationships (A < 0.05), the model fit improved overall and at the upper end, as shown by the solid black line (G = 0.79A–0.09, R2 = 0.51). These results are based on the assumption of no double reduction, i.e., that diploid gametes do not contain genes from sister chromatids; but in potato the probability of double reduction varies from 0 at the centromere to as high as 0.07 at the telomere (Bourke et al. 2015). In the context of a polygenic trait, with genes distributed across the entire chromosome, the effective value of the double reduction parameter is expected to be small. When the probability of double reduction was increased to 0.05 for computing A, the goodness of fit and intercept for the regression were unaffected but the slope decreased from 0.79 to 0.73.
Figure 1.
Comparison of off-diagonal elements for the additive covariance matrices estimated from markers (G) vs. pedigree (A) for the TP of 571 clones. The dashed line is the linear regression using all pairs of clones (G = 0.66A–0.06, R2 = 0.41) and the solid line is the regression (G = 0.79A–0.09, R2 = 0.51) when distant relationships (A < 0.05) are excluded.
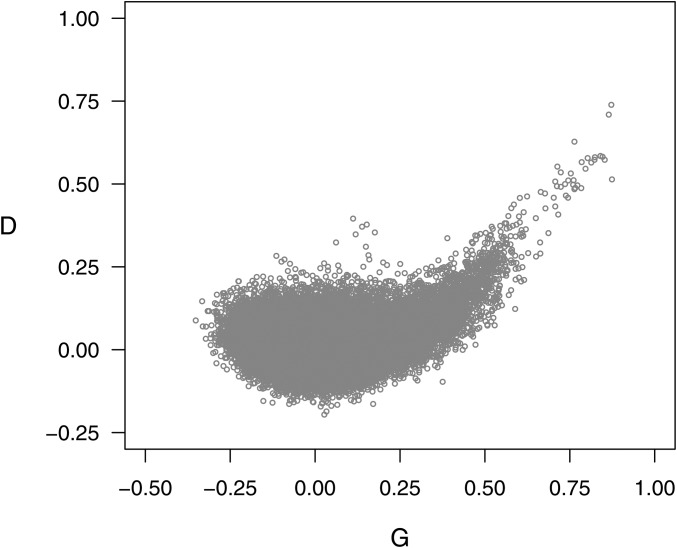
Figure 2 compares the off-diagonal elements of the digenic dominance covariance matrix D against the corresponding elements of G. For close relationships, Gij ≥ 0.4, G and D were highly correlated (r = 0.81); but when these pairs were excluded the correlation dropped to 0.08. The overall scaling of D is such that the mean of its diagonal elements equals 1 at panmictic equilibrium, which is very near the observed value of 0.99.
Figure 2.
Comparison of off-diagonal elements for the digenic dominance (D) vs. additive (G) covariance matrices estimated from markers for the TP of 571 clones.
A potential concern when using genomic relationship matrices is that estimates of genetic variance may be too low due to insufficient marker density. Yang et al. (2010) presented a method to assess (and correct) this issue. First, the markers are randomly partitioned, such that one half represent QTL and the other half markers; G (or D) is then calculated using each half separately; and finally GQTL is regressed onto Gmark. For both the G and D matrices in our data set, the mean regression coefficient after 100 iterations was 1.00 (SD 0.01), indicating sufficient marker density under the assumption that markers are sampled from the same distribution as QTL.
Variance components
Phenotype data for three economically important traits were analyzed: total yield, specific gravity (as a proxy for dry matter content), and potato chip fry color after 6 months of storage. Initially, genotype effects were modeled as independent to estimate the total genetic variance (Vg) and the variance of the genotype × year interaction (Vgy). The Vg estimate was higher than Vgy for all traits, ranging from 2.7× higher for yield to 5.6× higher for specific gravity (Table 3).
Table 3. Variance parameter estimates for the total genotypic value and genotype × year effect.
| Variance component | Yield (Mg2 ha−2) | Specific gravity | Fry color (L*2) |
|---|---|---|---|
| Vg | 88 | 26.5 × 10−6 | 5.1 |
| Vgy | 33 | 4.7 × 10−6 | 1.4 |
Vg, total genotypic value; Vgy, genotype × year effect.
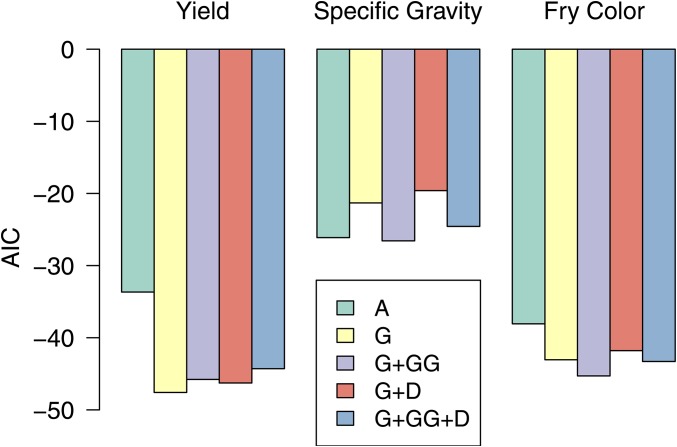
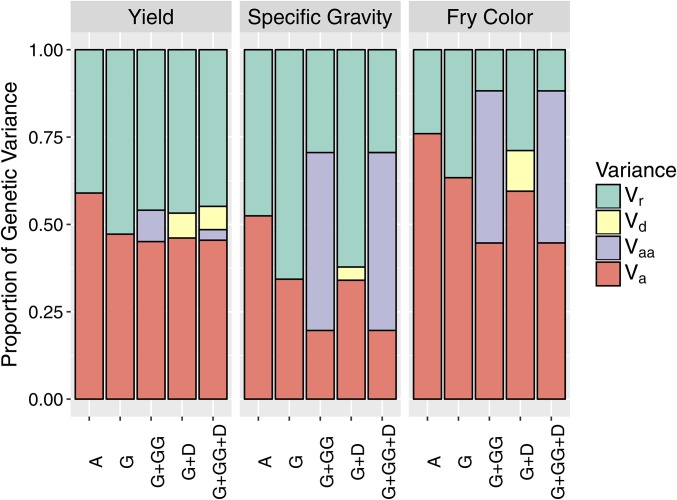
By adding another random effect to the baseline model, with covariance proportional to A or G, the genetic variance (Vg) was partitioned into additive (Va) and residual genetic variance (Vr), the latter corresponding to the independent clone effect (Table 1). Both A and G lowered the AIC compared to the baseline model for all traits, with the G matrix producing a better fit for yield and fry color vs. the A matrix for specific gravity (Figure 3). Using A, the proportion of genetic variance due to additive effects was 0.52 for specific gravity, 0.59 for yield, and 0.76 for fry color (Figure 4). When G was used, the additive genetic variance estimates were reduced by 0.12–0.18 of the total genetic variance, depending on the trait.
Figure 3.
Comparing goodness of fit in the TP for different genetic covariance structures (see Table 1) using the AIC relative to a baseline model with independent clone effects.
Figure 4.
Partitioning of genetic variance in the TP of 571 clones for five different covariance structures (see Table 1). Va, additive variance; Vaa, additive × additive epistasis; Vd, digenic dominance; Vr, residual genetic variance (for independent clone effects).
For specific gravity and fry color, including additive × additive epistasis lowered the AIC compared to the additive G-BLUP model; but the AIC increased when dominance was included (Figure 3). For these traits, a substantial amount of the estimated additive variance in the G model became additive × additive epistasis in the G+GG model: Va dropped from 34 to 20% of Vg for specific gravity and from 63 to 45% for fry color, with 44–51% of the genetic variance captured by G#G (Figure 4, SE in Table S2 in File S1). For yield, neither dominance nor additive × additive epistasis improved the AIC compared to G-BLUP, and only 10% of the genetic variance was captured by D or G#G compared to 45% for the residual clone effect.
Prediction accuracy
Many studies use random cross-validation to assess the accuracy of genome-wide prediction. However, in the context of a pedigreed breeding population, this approach leads to training-set individuals that are descendants of individuals in the validation set, which is unrepresentative of how GS will be used in practice and may produce unrealistically high accuracies. To avoid this pitfall, we used the pedigree depth metric to partition the population into a set of 168 candidates for selection (depth ≥12) and a set of 403 clones ancestral to this group (depth <12) as the training set. The selection candidates were further narrowed by excluding clones with insufficiently reliable data for validation, leaving 54 clones for yield, 132 clones for specific gravity, and 49 clones for fry color (with mean reliability in the range 0.71–0.72 for all traits).
Figure 5 shows the accuracy (left) and regression coefficient (right) when using each of the models in Table 1 to predict total genotypic value in the validation set. Prediction accuracy using only the A matrix was just over 0.5 for total yield vs. 0.4 for specific gravity and fry color. Replacing A with G improved accuracy for yield by 0.03 and fry color by 0.06, but decreased the accuracy for specific gravity by 0.07, which is consistent with the trend observed for AIC. Including dominance improved yield accuracy by 0.01 and including epistasis improved specific gravity accuracy by 0.05. Based on the AIC results for fry color, we expected higher accuracy with the G+GG model, but this was not observed. Including epistasis reduced prediction bias for fry color and specific gravity, with regression coefficients >0.9 for the G+GG model compared to 0.75–0.78 for G-BLUP.
Figure 5.
Prediction accuracy (left) and bias (right) when using all clones with pedigree depth <12 to predict clones with pedigree depth ≥12. SpGr, specific gravity.
To investigate the effect of training population size on accuracy, 200 random subsets of the TP were taken at N = 100, 200, and 300 clones (Figure S3 in File S1). For all three traits, prediction accuracy decreased as the TP was reduced. Fry color accuracy was the most sensitive, dropping by 0.28 when population size was reduced from 403 to 100, compared to accuracy decreases of 0.14 and 0.19 for yield and specific gravity, respectively.
We also determined accuracy when using the entire (N = 571) TP to predict yield and specific gravity in six unselected F1 populations (Table 4). The F1 populations ranged in size from 48 to 167 clones, and the number of polymorphic markers ranged from 1376 to 2311 (Table 2). Several of the parents had little pedigree relationship to the TP because they were russet clones, which is a distinct market category from the round, chip-processing type. Prediction accuracies ranged from 0.06 to 0.63 with G-BLUP, with no discernible connection between accuracy and pedigree relationship to the TP. The G+GG+D model performed very similarly, with no difference in average accuracy (to two decimal places) across the eight cases in Table 4. To assess the value of tetraploid allele dosage, predictions were made with “diploidized” marker data (Gdip) in which the three heterozygotes were recoded to be identical. The accuracy of the Gdip model was consistently lower, with an average loss of 0.13.
Table 4. Prediction accuracy in unselected F1 populations.
| Population | Trait (reliabilitya) | G model accuracy | G+GG+D accuracy | Gdip accuracy |
|---|---|---|---|---|
| W12011 | SpGr (0.74) | 0.63 | 0.61 | 0.36 |
| W12012 | Yield (0.66) | 0.31 | 0.31 | 0.06 |
| W12012 | SpGr (0.70) | 0.27 | 0.32 | 0.16 |
| W12060 | SpGr (0.84) | 0.25 | 0.29 | 0.13 |
| W9817 | Yield (0.82) | 0.06 | 0.06 | 0.08 |
| W10010 | Yield (0.82) | 0.12 | 0.11 | 0.14 |
| W×L | Yield (0.65) | 0.34 | 0.34 | 0.16 |
| W×L | SpGr (0.85) | 0.33 | 0.33 | 0.19 |
Gdip, G matrix based on diploidized marker data; SpGr, specific gravity.
Mean reliability of the validation data.
Discussion
This is the first study to connect the classical theory of genetic variance partitioning in tetraploids with covariance matrices constructed from genome-wide allele dosage information. Our results are based on a two-stage analysis in which the genotype estimate for each clone × environment combination was calculated in stage one by assuming independent effects, and in stage-two genomic covariance matrices were used. We note that not all references to two-stage analysis in the literature employ this convention; often a single genotype mean across all environments is estimated in the first stage. In our data set, there were 849 genotype × year means estimated in stage one for the 571 clones (File S4), and this partial replication is expected to improve the precision of the variance component estimates compared to analyzing a single mean per genotype in stage two (Kruijer et al. 2015).
The partial replication across years also allowed for explicit modeling of a residual genetic effect with no covariance structure in addition to the additive, dominance, and epistatic random effects. We interpret the residual genetic effect to include higher-order nonadditive effects (e.g., trigenic dominance, additive × dominance epistasis) and genetic variance not captured by the markers. The latter might appear to be ruled out based on our analysis of the G and D matrices using the Yang et al. (2010) method, but this assumes QTL are drawn from the same distribution as the markers. In reality, low-frequency alleles are underrepresented on the potato 8303 SNP array and are expected to contribute residual genetic variance (Vos et al. 2015). The estimates for Vd and Vaa were sensitive to whether the residual genetic effect was included in the model. Without it, Vaa for yield was estimated at 40 (SE 14) Mg2 ha−2, which is 45% of the total genetic variance (88 Mg2 ha−2). When the residual effect was included, most of this variance shifted into Vr. This phenomenon and the large SE of the estimates (Table S2 in File S1) suggest that the partitioning of the nonadditive genetic variance is uncertain, probably because of limited population size.
At first glance, our finding that more of the genetic variance was additive for yield (45%) compared to specific gravity (20%) seems unexpected. Diallel studies have typically found the ratio between SCA and GCA to be higher for yield. Tai (1976) reported SCA/GCA = 3.8 for yield vs. 0.6 for specific gravity, and Bradshaw et al. (2000) reported SCA/GCA = 0.75 for yield vs. 0.06 for specific gravity. Whereas these earlier studies used unselected populations, the clones in our TP had been selected for high specific gravity at least once, and in many cases for 2–3 years. Specific gravity (which is closely correlated with dry matter content) is one of the most important traits for the chip processing market, and strong selection early in the variety-development process is practiced because the trait shows relatively little genotype × environment interaction (Table 3; Wang et al. 2017). Our results suggest that, in the tail of the phenotype distribution for specific gravity, the partitioning of genetic variance is shifted toward nonadditive effects.
A unique feature of our study compared to previous reports of GS in autotetraploid species, such as alfalfa (Annicchiarico et al. 2015; Li et al. 2015) and potato (Habyarimana et al. 2017; Sverrisdóttir et al. 2017), was the use of SNP array data rather than genotyping by sequencing (GBS). A major benefit of the SNP array for polyploids is the ability to accurately determine allele dosage (Voorrips et al. 2011; Schmitz Carley et al. 2017), but the cost of the array is determined by sales volume and can be prohibitively expensive for GS in small breeding programs or minor crops. GBS methods achieve low per-sample costs by pooling many samples into one library for sequencing; but much higher read depth (per sample–marker combination) is needed for accurate genotype assignment in tetraploids compared to heterozygous diploids. By comparing GBS with KASP markers in potato, Uitdewilligen et al. (2013) recommended 60–80× read depth to differentiate the three heterozygous genotypes, which agrees well with theoretical calculations (J. B. Endelman, unpublished data). For GS, a pressing question is whether paying for more sequencing to improve estimates of allele dosage provides a return on investment in terms of prediction accuracy and ultimately genetic gain. A completely general answer may be elusive due to complex interactions between GBS method, population, and phenotype; but our finding that diploidization of the marker data consistently reduced prediction accuracy in the F1 populations (by 0.13 on average) highlights the need for further research.
In plant breeding, both the total genotypic value and additive value are relevant for selection. For many crops, the unit of commercial production (i.e., inbred, F1 hybrid, or vegetative clone) is the same genotype evaluated by the breeder and therefore selection should be based on total genotypic value. When selecting new parents, however, only the additive value should be considered because nonadditive effects are less efficiently transmitted to progeny. We have demonstrated the feasibility of this paradigm for potato using the G+GG+D model, although questions remain regarding its optimal implementation. For many issues, further progress will require larger populations genotyped with less ascertainment bias.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.118.300685/-/DC1.
Acknowledgments
Financial support was provided by Potatoes USA, the U.S. Department of Agriculture National Institute of Food and Agriculture, Award 2014-67013-22418, and U.S. Department of Agriculture Hatch Projects 1002731 and 1013047.
Author contributions: J.B.E. designed the study. J.B.E. and C.A.S.C. analyzed the data and drafted the manuscript. All authors contributed germplasm, data, or financial resources.
Footnotes
Communicating editor: M. Sillanpää
Literature Cited
- Amadeu R. R., Cellon C., Olmstead J. W., Garcia A. A., Resende M. F., et al. , 2016. AGHmatrix: R package to construct relationship matrices for autotetraploid and diploid species: a blueberry example. Plant Genome 9 10.3835/plantgenome2016.01.0009 [DOI] [PubMed] [Google Scholar]
- Annicchiarico P., Nazzicari N., Li X., Wei Y., Pecetti L., et al. , 2015. Accuracy of genomic selection for alfalfa biomass yield in different reference populations. BMC Genomics 16: 1020 10.1186/s12864-015-2212-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo R., 1994. Prediction of maize single-cross performance using RFLPs and information from related hybrids. Crop Sci. 34: 20–25. 10.2135/cropsci1994.0011183X003400010003x [DOI] [Google Scholar]
- Bourke P. M., Voorrips R. E., Visser R. G. F., Maliepaard C., 2015. The double-reduction landscape in tetraploid potato as revealed by a high-density linkage map. Genetics 201: 853–863. 10.1534/genetics.115.181008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw J. E., Todd D., Wilson R. N., 2000. Use of tuber progeny tests for genetical studies as part of a potato (Solanum tuberosum subsp. tuberosum) breeding programme. Theor. Appl. Genet. 100: 772–781. 10.1007/s001220051351 [DOI] [PubMed] [Google Scholar]
- Brown J., Caligari P. D. S., 1989. Cross prediction in a potato breeding programme by evaluation of parental material. Theor. Appl. Genet. 77: 246–252. 10.1007/BF00266194 [DOI] [PubMed] [Google Scholar]
- Butler D. G., Cullis B. R., Gilmour A. R., Gogel B. J., 2009. ASReml-R Reference Manual Version 3. Queensland Government Department of Primary Industries and Fisheries, Brisbane, Australia. [Google Scholar]
- Clark S. A., Hickey J. M., Daetwyler H. D., van der Werf J. H. J., 2012. The importance of information on relatives for the prediction of genomic breeding values and the implications for the makeup of reference data sets in livestock breeding schemes. Genet. Sel. Evol. 44: 4 10.1186/1297-9686-44-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damesa T. M., Möhring J., Worku M., Piepho H. P., 2017. One step at a time: stage-wise analysis of a series of experiments. Agron. J. 109: 845–857. 10.2134/agronj2016.07.0395 [DOI] [Google Scholar]
- da Silva W. L., Ingram J., Hackett C. A., Coombs J. J., Douches D., et al. , 2017. Mapping loci that control tuber and foliar symptoms caused by PVY in autotetraploid potato (Solanum tuberosum L.). G3 (Bethesda) 7: 3587–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers J. C. M., 2007. Prediction of response to marker-assisted and genomic selection using selection index theory. J. Anim. Breed. Genet. 124: 331–341. 10.1111/j.1439-0388.2007.00701.x [DOI] [PubMed] [Google Scholar]
- Felcher K. J., Coombs J. J., Massa A. N., Hansey C. N., Hamilton J. P., et al. , 2012. Integration of two diploid potato linkage maps with the potato genome sequence. PLoS One 7: e36347 10.1371/journal.pone.0036347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. A., 1941. Average excess and average effect of a gene substitution. Ann. Eugen. 11: 53–63. 10.1111/j.1469-1809.1941.tb02272.x [DOI] [Google Scholar]
- Frederick, C. M., 2017 Exploration of novel phenotyping techniques and identification of quantitative trait loci for chip processing potatoes. Ph.D. Thesis, University of Wisconsin, Madison. [Google Scholar]
- Gallais A., 2003. Quantitative Genetics and Breeding Methods in Autopolyploid Plants. Institut National de la Recherche Agronomique, Paris. [Google Scholar]
- Gopal J., 1998. General combining ability and its repeatability in early generations of potato breeding programmes. Potato Res. 41: 21–28. 10.1007/BF02360258 [DOI] [Google Scholar]
- Habier D., Fernando R. L., Dekkers J. C. M., 2007. The impact of genetic relationship information on genome-assisted breeding values. Genetics 177: 2389–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habyarimana E., Parisi B., Mandolino G., 2017. Genomic prediction for yields, processing and nutritional quality traits in cultivated potato (Solanum tuberosum L.). Plant Breed. 136: 245–252. 10.1111/pbr.12461 [DOI] [Google Scholar]
- Hamilton J. P., Hansey C. N., Whitty B. R., Stoffel K., Massa A. N., et al. , 2011. Single nucleotide polymorphism discovery in elite North American potato germplasm. BMC Genomics 12: 302 10.1186/1471-2164-12-302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes B. J., Visscher P. M., Goddard M. E., 2009. Increased accuracy of artificial selection by using the realized relationship matrix. Genet. Res. 91: 47–60. 10.1017/S0016672308009981 [DOI] [PubMed] [Google Scholar]
- Henderson C. R., 1975. Best linear unbiased estimation and prediction under a selection model. Biometrics 31: 423–447. 10.2307/2529430 [DOI] [PubMed] [Google Scholar]
- Henderson C. R., 1985. Best linear unbiased prediction of non-additive genetic merits in noninbred populations. J. Anim. Sci. 60: 111–117. 10.2527/jas1985.601111x [DOI] [Google Scholar]
- Jiang Y., Reif J. C., 2015. Modeling epistasis in genomic selection. Genetics 201: 759–768. 10.1534/genetics.115.177907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempthorne O., 1955a The theoretical values of correlations between relatives in random mating populations. Genetics 40: 153–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempthorne O., 1955b The correlation between relatives in a simple autotetraploid population. Genetics 40: 168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempthorne O., 1955c The correlations between relatives in random mating populations. Cold Spring Harb. Symp. Quant. Biol. 20: 60–78. 10.1101/SQB.1955.020.01.008 [DOI] [PubMed] [Google Scholar]
- Kempthorne O., 1957. An Introduction to Genetic Statistics. John Wiley & Sons, New York. [Google Scholar]
- Kerr R. J., Li L., Tier B., Dutkowski G. W., McRae T. A., 2012. Use of the numerator relationship matrix in genetic analysis in autopolyploid species. Theor. Appl. Genet. 124: 1271–1282. 10.1007/s00122-012-1785-y [DOI] [PubMed] [Google Scholar]
- Kruijer W., Boer M. P., Malosetti M., Flood P. J., Engel B., et al. , 2015. Marker-based estimation of heritability in immortal populations. Genetics 199: 379–398. 10.1534/genetics.114.167916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legarra A., 2016. Comparing estimates of genetic variance across different relationship models. Theor. Popul. Biol. 107: 26–30. 10.1016/j.tpb.2015.08.005 [DOI] [PubMed] [Google Scholar]
- Li X., Wei Y., Acharya A., Hansen J. L., Crawford J. L., et al. , 2015. Genomic prediction of biomass yield in two selection cycles of a tetraploid alfalfa breeding population. Plant Genome 8 10.3835/plantgenome2014.12.0090 [DOI] [PubMed] [Google Scholar]
- Lopez-Pardo R., Barandalla L., Ritter E., Ruiz De Galarreta J. I., 2013. Validation of molecular markers for pathogen resistance in potato. Plant Breed. 132: 246–251. 10.1111/pbr.12062 [DOI] [Google Scholar]
- Maris B., 1989. Analysis of an incomplete diallel cross among three ssp. tuberosum varieties and seven long-day adapted ssp. andigena clones of the potato (Solanum tuberosum L.). Euphytica 41: 163–182. 10.1007/BF00022425 [DOI] [Google Scholar]
- Meuwissen T. H. E., Hayes B. J., Goddard M. E., 2001. Prediction of the total genetic value using genome-wide dense marker maps. Genetics 157: 1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz P. R., Resende M. F. R., Gezan S. A., Resende M. D. V., de los Campos G., et al. , 2014. Unraveling additive from nonadditive effects using genomic relationship matrices. Genetics 198: 1759–1768. 10.1534/genetics.114.171322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neele A. E. F., Nab H. J., Louwes K. M., 1991. Identification of superior parents in a potato breeding programme. Theor. Appl. Genet. 82: 264–272. 10.1007/BF02190611 [DOI] [PubMed] [Google Scholar]
- Nejati-Javaremi A., Smith C., Gibson J. P., 1997. Effect of total allelic relationship on accuracy of evaluation and response to selection. J. Anim. Sci. 75: 1738–1745. 10.2527/1997.7571738x [DOI] [PubMed] [Google Scholar]
- Ould Estaghvirou S. B., Ogutu J. O., Schulz-Streeck T., Knaak C., Ouzunova M., et al. , 2013. Evaluation of approaches for estimating the accuracy of genomic prediction in plant breeding. BMC Genomics 14: 860 10.1186/1471-2164-14-860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepho H. P., 2009. Ridge regression and extensions for genomewide selection in maize. Crop Sci. 49: 1165–1176. 10.2135/cropsci2008.10.0595 [DOI] [Google Scholar]
- Plaisted R. L., Sanford L., Federer W. T., Kehr A. E., Peterson L. C., 1962. Specific and general combining ability for yield in potatoes. Am. Potato J. 39: 185–197. 10.1007/BF02871402 [DOI] [Google Scholar]
- Rak K., Palta J. P., 2015. Influence of mating structure on agronomic performance, chip fry color, and genetic distance among biparental tetraploid families. Am. J. Potato Res. 92: 518–535. 10.1007/s12230-015-9466-4 [DOI] [Google Scholar]
- R Development Core Team , 2015. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. [Google Scholar]
- Schmitz Carley C. A., Coombs J. J., Douches D. S., Bethke P. C., Palta J. P., et al. , 2017. Automated tetraploid genotype calling by hierarchical clustering. Theor. Appl. Genet. 130: 717–726. 10.1007/s00122-016-2845-5 [DOI] [PubMed] [Google Scholar]
- Slater A. T., Wilson G. M., Cogan N. O. I., Forster J. W., Hayes B. J., 2014. Improving the analysis of low heritability complex traits for enhanced genetic gain in potato. Theor. Appl. Genet. 127: 809–820. 10.1007/s00122-013-2258-7 [DOI] [PubMed] [Google Scholar]
- Slater A. T., Cogan N. O., Forster J. W., Hayes B. J., Daetwyler H. D., 2016. Improving genetic gain with genomic selection in autotetraploid potato. Plant Genome 9 10.3835/plantgenome2016.02.0021 [DOI] [PubMed] [Google Scholar]
- Smith A. B., Cullis B. R., Gilmour A., 2001. The analysis of crop variety evaluation data in Australia. Aust. N. Z. J. Stat. 43: 129–145. 10.1111/1467-842X.00163 [DOI] [Google Scholar]
- Su G., Christensen O. F., Ostersen T., Henryon M., Lund M. S., 2012. Estimating additive and non-additive genetic variances and predicting genetic merits using genome-wide dense single nucleotide polymorphism markers. PLoS One 7: e45293 10.1371/journal.pone.0045293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sverrisdóttir E., Byrne S., Sundmark H. E. R., Johnsen H. Ø., Kirk H. G., et al. , 2017. Genomic prediction of starch content and chipping quality in tetraploid potato using genotyping-by-sequencing. Theor. Appl. Genet. 130: 2091–2108. 10.1007/s00122-017-2944-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai G. C. C., 1976. Estimation of general and specific combining abilities in potato. Can. J. Genet. Cytol. 18: 463–470. 10.1139/g76-056 [DOI] [Google Scholar]
- Uitdewilligen J. G., Wolters A. M., D’hoop B. B., Borm T. J., Visser R. G., et al. , 2013. A next-generation sequencing method for genotyping-by-sequencing of highly heterozygous autotetraploid potato. PLoS One 8: e62355 (erratum: PLoS One 10: e0141940) 10.1371/journal.pone.0062355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berloo R., Hutten R. C. B., van Eck H. J., Visser R. G. F., 2007. An online potato pedigree database resource. Potato Res. 50: 45–57. 10.1007/s11540-007-9028-3 [DOI] [Google Scholar]
- VanRaden P. M., 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91: 4414–4423. 10.3168/jds.2007-0980 [DOI] [PubMed] [Google Scholar]
- Vitezica Z. G., Varona L., Legarra A., 2013. On the additive and dominant variance and covariance of individuals within the genomic selection scope. Genetics 195: 1223–1230. 10.1534/genetics.113.155176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorrips R. E., Gort G., Vosman B., 2011. Genotype calling in tetraploid species from bi-allelic marker data using mixture models. BMC Bioinformatics 12: 172 10.1186/1471-2105-12-172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos P. G., Uitdewilligen J. G. A. M. L., Voorrips R. E., Visser R. G. F., van Eck H. J., 2015. Development and analysis of a 20K SNP array for potato (Solanum tuberosum): an insight into the breeding history. Theor. Appl. Genet. 128: 2387–2401. 10.1007/s00122-015-2593-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Snodgrass L. B., Bethke P. C., Bussan A. J., Holm D. G., et al. , 2017. Reliability of measurement and genotype x environment interaction for potato specific gravity. Crop Sci. 57: 1966–1972. 10.2135/cropsci2016.12.0976 [DOI] [Google Scholar]
- Wientjes Y. C. J., Bijma P., Vandenplas J., Calus M. P. L., 2017. Multi-population genomic relationships for estimating current genetic variances within and genetic correlations between populations. Genetics 207: 503–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wricke G., Weber W. E., 1986. Quantitative Genetics and Selection in Plant Breeding. Walter de Gruyter & Co., Berlin: 10.1515/9783110837520 [DOI] [Google Scholar]
- Xu S., 2013. Mapping quantitative trait loci by controlling polygenic background effects. Genetics 195: 1209–1222. 10.1534/genetics.113.157032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Benyamin B., McEvoy B. P., Gordon S., Henders A. K., et al. , 2010. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42: 565–569. 10.1038/ng.608 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All marker, pedigree, and phenotype data needed to reproduce the results in this manuscript are provided in File S2, File S3, File S4, File S5, and File S6: