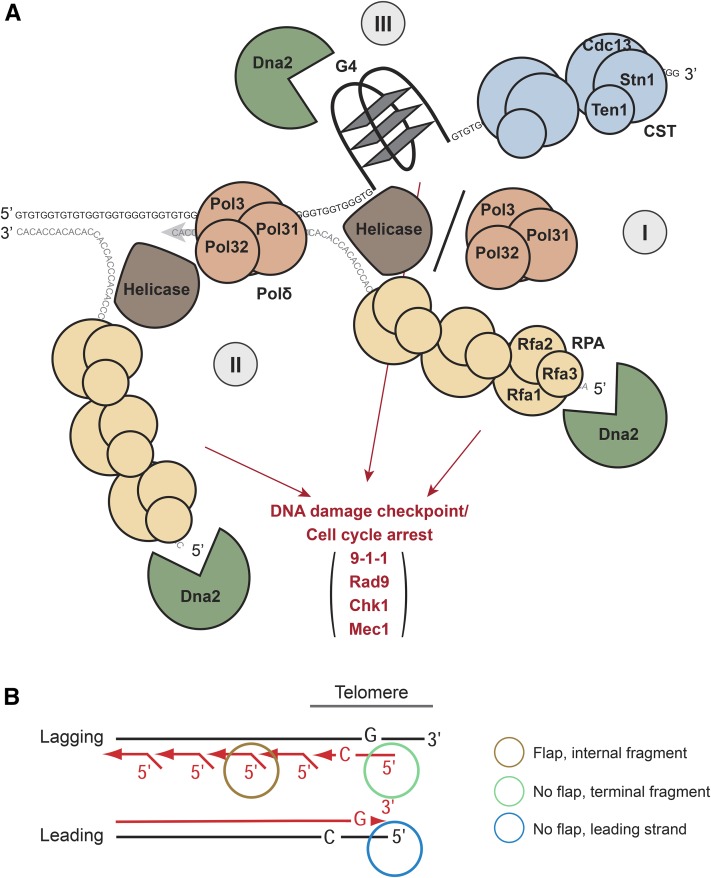
Figure 6.
Three plausible roles for Dna2 in removing unwound RPA-coated ssDNA at telomeres. (A) Three scenarios for Dna2 activity. Scenario I: 5′ RPA-coated ssDNA cleavage at telomeric termini. Telomere ends are unwound by helicases, for example, Pif1 or Mph1. The 3′ G-rich strand is bound by CST and the 5′ C-rich strand is bound by RPA, a substrate for Dna2 cleavage. Scenario II: Processing of long flaps on Okazaki fragments near telomeres. DNA polymerase δ displacement activity, stimulated by helicase(s), generates long flaps on an Okazaki fragment near telomere. Long C-rich flap, bound by RPA, are subjected to Dna2 cleavage. Scenario III: G-quadruplex unwinding and processing. G-quadruplexes formed on telomeric G-rich ssDNA are unwound or processed by Dna2. All proteins were drawn to scale. (B) Lagging and leading strand replication at telomeres. Short red arrows indicate Okazaki fragments on the lagging strand. The long red arrow indicates replicated leading strand. The brown circle indicates the flap formed on an internal Okazaki fragment. The green circle indicates no flap on the terminal telomeric Okazaki fragment. The blue circle indicates no flap on the leading strand template.