Figure 3. Brd4 is dispensable in naïve ESCs.
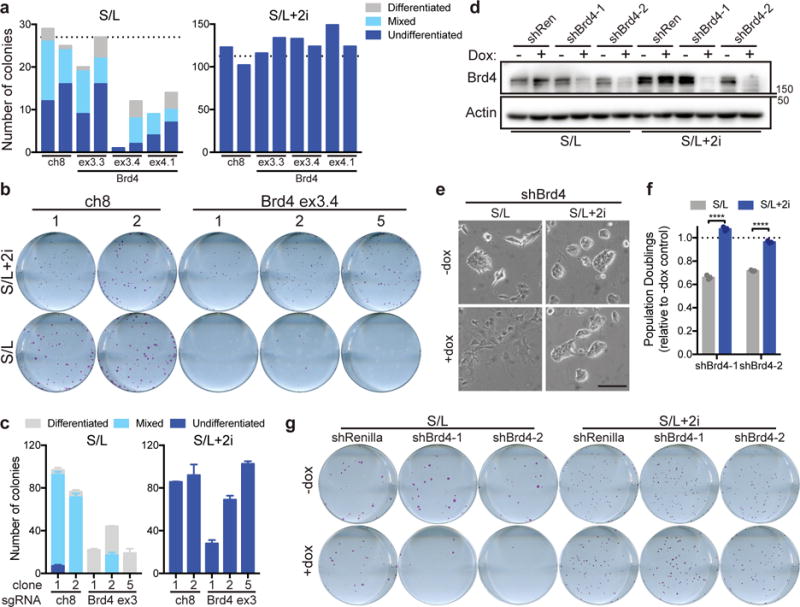
(a) Quantification of colony formation assay of cells cultured in S/L (left) or S/L+2i (right) transfected with Cas9 and the indicated sgRNA against a nongenic region of chromosome 8 (ch8, control) or exon 3 (ex3) or exon 4 (ex4) of Brd4. Dotted line represents average of ch8 controls. Each bar represents quantification of a single well of a six-well plate; transfected samples were seeded in duplicate. (b) Alkaline phosphatase staining of colonies formed from single cells of clonal ESC lines edited with sgRNA against chromosome 8 (ch8) or Brd4 exon 3 and cultured in S/L or S/L+2i. One representative well of a six-well plate is shown. (c) Quantification of colony formation assay shown in (b). (d) Western blot depicting Brd4 levels in ESCs expressing doxycycline (dox)-inducible hairpins against Renilla (shRen) or Brd4 (shBrd4). Cells were cultured with or without dox for 48 h prior to harvest. Actin is used as a loading control. Western blot was performed two independent times. (e) Brightfield images of ESCs expressing shBrd4-2 cultured for 48 h with or without doxycycline (dox). (f) Population doublings of cells cultured for 72 h in doxycycline relative to controls grown without dox. (g) Alkaline phosphatase staining of colonies formed from single cells expressing the indicated hairpins grown in the presence or absence of doxycycline (dox) and cultured in S/L or S/L+2i. One representative well of a six-well plate is shown. All bars represent mean ±SEM (c) or ±SD (f) of n=3 independent samples. ****, P < 0.0001 by 2-way ANOVA with Sidak’s multiple comparisons post test (shBrd4-1, P = 7.2e-10; shBrd4-2, P = 4.6e-8). Scale bar, 100 μm.
